文章信息
- 刘雪莹, 王广策, 张宝玉, 宫相忠. 2022.
- LIU Xue-ying, WANG Guang-ce , ZHANG Bao-yu, GONG Xiang-zhong. 2022.
- 条斑紫菜丙酮酸羧化酶基因表达对不同碳源的响应
- Expression of the pyruvate carboxylase gene in Pyropia yezoensis in response to different inorganic carbon sources
- 海洋科学, 46(12): 138-147
- Marine Sciences, 46(12): 138-147.
- http://dx.doi.org/10.11759/hykx20220422001
-
文章历史
- 收稿日期:2022-04-22
- 修回日期:2022-07-14
2. 中国科学院海洋研究所 实验海洋生物学重点实验室, 山东 青岛 266071;
3. 青岛海洋科学与技术试点国家实验室 海洋生物学与生物技术功能实验室, 山东 青岛 266237;
4. 中国科学院海洋大科学研究中心, 山东 青岛 266071
2. Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China;
3. Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266237, China;
4. Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
条斑紫菜(Pyropia yezoensis)是属于原红藻纲(Protoflorideae)、红毛菜目(Bangiales)、红毛菜科(Bangiaceae)、紫菜属(Pyropia)的一种大型海藻, 是东南亚地区重要的经济海藻之一。它的生活史是由丝状孢子体和叶状配子体组成, 属于异型世代交替[1-2]。紫菜营养丰富, 富含氨基酸、蛋白质、不饱和脂肪酸及多种微量元素, 是低脂肪高蛋白的健康绿色食品; 因其含有丰富的多糖尤其是琼胶, 也使其成为重要的化工原料; 紫菜除了经济价值和营养价值外, 其叶状体在生长过程中可大量吸收C、N、P等, 是海洋生态系统中重要的碳库、氮库和磷库[3]。紫菜作为潮间带海藻, 野生型叶状体随潮汐变化会经历暴露在空气中数小时和没入水中的生境变化, 在规模化养殖过程中, 渔民也需要定期将紫菜苗帘暴露在空气中以去除杂藻及提高品质[4]。为更深入了解紫菜, 国内外学者对紫菜的生活史、影响生长因素、抗逆和代谢产物等方面开展了大量的研究, 使之逐渐成为研究红藻的代表性物种[5-7]。
丙酮酸羧化酶(Pyruvate carboxylase, PYC)最早在鸡的肝脏线粒体中发现[8], 后来在非光合生物真菌线粒体中也发现了PYC。它属于生物素酶家族, 其特征是具有共价连接的生物素作为辅助因子, 依赖ATP, 催化丙酮酸和HCO3−生成草酰乙酸(oxaloacetate, OAA), 该反应作为糖异生的第一步, 在动物维持代谢稳态中具有重要作用[9]。如哺乳动物中PYC的缺失会引发神经系统紊乱, 代谢性酸中毒或癌症等多种代谢疾病[10-11]。缺乏PYC基因的酿酒酵母(Saccharomyces cerevisiae)突变体在葡萄糖作为唯一碳源时, 不能存活[12-13]。1990年, 研究者在胡萝卜、玉米、向日葵和油菜的细胞提取液发现了PYC活性, 首次证明了植物中PYC的存在, 但尚未明确其在植物不同部位或发育过程中的功能[14]。随着研究技术的发展和水平的提高, PYC陆续被发现存在微藻中[15-16]。Tsuji等[17]证实丙酮酸羧化酶存在海洋单细胞藻Emiliania huxleyi质体中, Km(米氏常数: 酶促反应达到最大反应速度一半时对应的底物浓度)值显示该质体丙酮酸羧化酶对丙酮酸有较高的亲和性, 主动参与羧化用于回补OAA, 作者推测PYC或许在各种水生光合生物中起着不可或缺的作用。当绿藻Coccomyxa subellipsoidea C-169培养在2% CO2环境时, PYC的RNA水平和酶活均增加, 作者认为PYC参与CO2固定进而提高C. subellipsoidea无机碳利用率[16]。综上所述, PYC似乎在促进水生光合生物无机碳利用方面发挥一定的作用, 到目前为止我们对此还是了解较少。我们前期在分析条斑紫菜叶状体和丝状体对不同无机碳响应的转录组数据(该转录组数据见国家基因库
以条斑紫菜叶状体cDNA为模板扩增获得PyPYC基因全长编码区序列后, 与来自其他物种的丙酮酸羧化酶核酸及蛋白序列进行比对分析, 明确PyPYC序列特征。同时用加富CO2的空气或改变人工海水中NaHCO3浓度方式设置不同无机碳源条件培养紫菜叶状体, 探究PYC基因在条斑紫菜叶状体无机碳利用中的响应。
1 材料与方法 1.1 材料条斑紫菜叶状体来自中国科学院海洋研究所藻类种质库, 在含有PES营养盐的无菌海水中培养。培养条件为: 温度15~18 ℃, 光照强度15~30 µmol·m−2·s−1, 光暗周期为14 h∶10 h, 每周更换1次培养基。
1.2 方法 1.2.1 条斑紫菜不同碳源处理选取叶片大小相近且生长旺盛的健康藻体为材料, 分别置于含不同浓度NaHCO3人工海水培养, NaHCO3终浓度分别为2、4、8 mmol/L。NaHCO3浓度设置基于在15 ℃, pH 8.07时, 自然海水中可溶性无机碳浓度为2.1 mmol/L, 而可溶性无机碳的主要成分是HCO3−, 占91%[18]。每150 mL人工海水培养0.03 g新鲜藻体, 其余培养条件同上。参考文献[19-20]室内培养紫菜方法, 每2 d全部更换培养基的培养条件, 我们将培养在不同NaHCO3浓度的叶状体培养24 h后, 光照时采收。
在以CO2为碳源的处理中, 我们将条斑紫菜叶状体平铺于细胞培养瓶中的滤纸上并一直保持湿润, 分别往培养瓶中通入正常空气(CO2体积分数为4.17×10-4)和加富CO2的空气(CO2终体积分数分别为1×10-3和2×10-3)。青岛附近海域的半日潮使得野外生长的条斑紫菜干出时间约为2~4 h, 因此我们将条斑紫菜叶状体处理3 h后采收, 吸干表面水分后用液氮冻存, 以备后续使用。在处理过程中始终保持不低于60%的含水量, 光照强度30 µmol·m−2·s−1, 温度15~18 ℃。以上实验设计每组处理设置3个平行。
1.2.2 RNA提取及反转录制备cDNA条斑紫菜叶状体RNA提取及反转录方法参考文献[21]。简介如下: 约100 mg新鲜培养的叶状体用吸水纸吸掉表面的水分, 迅速用液氮冷冻研磨, 然后用植物RNA提取试剂盒(天根, 北京)提取。提取的RNA首先用1%的琼脂糖凝胶电泳检验其质量, 然后用Nanodrop Photometer分光光度计(德国)测定RNA的纯度与浓度。
用于扩增目的基因的cDNA采用MMLV试剂盒(promega)反转, 反转过程未去除基因组DNA。用于实时荧光定量PCR(RT-qPCR)的cDNA依据Takara反转录酶试剂盒说明书(TaKaRa, 大连)进行, 具体实验操作步骤如下: 1) 去除基因组DNA反应。5×gDNA Eraser Buffer 2 μL, gDNA Eraser 1μL, Total RNA (1 000 ng), 添加RNase Free dH2O至10 μL; 42℃ 2 min; 2) 反转录反应。向步骤1的反应液中加入以下试剂, PrimeScript RT Enzyme Mix I 1 μL, RT Primer Mix 1 μL, 5×Primescript Buffer 4 μL, RNase Free dH2O 4 μL; 37 ℃ 30 min, 85 ℃ 5 s, 4 ℃保存。所获cDNA模板置于−20 ℃暂存。
1.2.3 目的基因扩增分析转录组unigene TRINITY_DN105351_c0_g和TRINITY_DN15039_c0_g1序列后, 利用Primer Premier 5.0设计用于扩增目的基因的引物, 引物名称及序列见表 1。采用Touchdown程序, 即: 94 ℃ 2 min, 98 ℃ 30 s, 接下来10循环98 ℃ 30 s, 65 ℃ 15 s, 68 ℃ 150 s, 每个循环退火温度降低0.1 ℃, 接下来进入30个循环为98 ℃ 30 s, 55 ℃ 15 s, 68 ℃ 150 s, DNA聚合酶为Tks Gflex (TaKaRa, 大连), 所用仪器为SensoQuest Labcycler (SensoQuest, 德国)。
| 引物名称 | 序列(5′—3′) |
| PyPYC1-F | ATGTATTTACGCGCAAAGTCAGTAAACT |
| PyPYC1-R | TTATTCCAGTTCTACTATCAAGTCTCCAGCT |
| PyPYC2-F | ATGTTTGGCACCAAGACGTCG |
| PyPYC2-R | TCACGCCCCAGCCGC |
| qPYC-F | CTACCGCAACGCTGGCACTG |
| qPYC-R | CTCTTCGCTCACCGTGTGTTCC |
| GAPDH-F | CCAACAAGTGGGAGTAAGCG |
| GAPDH-R | GGACAGAACCGAACAGCGTA |
所获得的序列在NCBI (
用于RT-qPCR的引物名称及序列见表 1, 并在图 1B中用双下划线标出。以GAPDH为内参基因[25]。反应所用试剂购自Roche公司(Switzerland), 仪器为Quant Studio1 (Thermo Fisher Scientific Inc., U. S.)。PCR反应条件为: 94 ℃ 2 min, 接下来40循环为94 ℃ 15 s, 60 ℃ 15 s, 72 ℃ 20 s, 扩增结束后的溶解曲线分析为检测扩增产物特异性。RT-qPCR数据采用相对定量法分析, 用SPSS单因素方差分析检验显著性差异水平。
引物对PyPYC1-F/PyPYC1-R和PyPYC2-F/ PyPYC2-R分别对应TRINITY_DN15039_c0_g1和TRINITY_DN105351_c0_g1序列。尽管在模板浓度、退火温度以及Taq酶等方面都做了很多尝试, 用PyPYC1-F/PyPYC1-R引物无法获得PCR产物, 也就是说不能获得RINITY_DN15039_c0_g1对应序列。而且, 该unigene序列在条斑紫菜基因组序列[7] (BioProject PRJNA589917 in NCBI)中也无匹配序列。因此, 我们初步认为来自转录组的TRINITY_ DN15039_c0_g1为来自细菌污染。采用引物对PyPYC2-F和PyPYC2-R获得大约4 K的PCR产物(图 1a)。测序后共获得3 546 bp的核酸序列, 除去324 bp的内含子区外, 其余部分与TRINITY_ DN105351_c0_g1相同(图 1b)。该序列对应条斑紫菜基因组第一条染色体上从14 819 602至14 823 147区域的核酸序列。经BlastX比对发现, 该序列与来自红藻Porphyridium purpureum PYC1氨基酸序列(序列入口号: KAA8491876.1)有61%的相似性, 与细菌Sandaracinaceae bacterium的PYC氨基酸序列(RZO64306.1)有53%的相似性, 从而确定该产物为条斑紫菜的PYC基因, 命名为PyPYC。
2.2 序列特征分析及系统进化树分析丙酮酸羧化酶催化丙酮酸和HCO3−结合生成草酰乙酸的整个反应分为以下两步进行[26-28]:
| $\text { Enz-biotin }+\mathrm{ATP}+\mathrm{HCO}_3^{-} \stackrel{\text { Acetyl-CoA } \mathrm{Mg}^{2+}}{\longleftrightarrow} \mathrm{ADP}+\mathrm{Pi}+\text { Enz-biotin- } \mathrm{CO}_2^{-} $ | (1) |
| $ \text { Enz-biotin- } \mathrm{CO}_2^{-}+\text {pyruvate } \longrightarrow \text { Enz-biotin }+\text { oxaloacetate } $ | (2) |
在结构上, 典型的丙酮酸羧化酶由4个相同的亚基排列成四面体状结构, 每个亚基包含3个功能结构域: 生物素羧化(BC, biotin carboxylase)结构域、转羧基(CT, carboxyltransferase) 结构域以及生物素羧基载体(BCCP, biotin carboxyl carrier protein)结构域[27, 29]。这种依赖生物素的羧化酶在2个活性位点以2个连续的步骤执行它们的活性, 在第一步反应中, 可移动的羧基载体生物素在BC活性部位进行羧化, 这一过程需要Mg-ATP提供能量。然后, 结合了羧基的生物素被BCCP转移到CT活性中心将羧基转移到底物丙酮酸上, 使丙酮酸完成羧化反应[30]。并且, PYC的每个亚基都含有一个紧密结合的二价金属离子(Zn2+/Mg2+), 它作为PYC的辅助因子似乎起着结构性的作用[27]。
我们采用ClustalX软件将不同来源的PYC氨基酸序列进行比对和相似性分析(表 2和图 2), 发现来自不同物种的PYC氨基酸序列具有较高的保守性, PyPYC与来自Coccomyxa subellipsoidea C-169、Saccharomyces cerevisiae和Rhizobium phaseoli Ch24-10的PYC分别有41.2%, 42.6%和38.3%相似性(表 2)。在这些物种PYC序列上已经证实的活性位点区域, 如与Mg-ATP结合的位点(ERNCSIQRRHQKV和QVEH), BCCP结构域中的生物素结合位点(AMKM), CT结构域中丙酮酸结合位点(ETWGGATFDVAM RFLECPWERL)和1个二价金属离子(Zn2+/Mg2+)结合位点(HVHTH)等[31], 在PyPYC序列中也同样存在, 且序列相似度较高(图 2)。
| 序列 | PyPYC | CsPYC | ScPYC | RpPYC |
| PyPYC | 1 | |||
| CsPYC | 0.412 | 1 | ||
| ScPYC | 0.426 | 0.468 | 1 | |
| RpPYC | 0.383 | 0.423 | 0.440 | 1 |
| 注: Cs代表Coccomyxa subellipsoidea C-169; Sc代表Sac-cha-ro-myces cerevisiae; Rp代表 Rhizobium phaseoli Ch24-10 | ||||
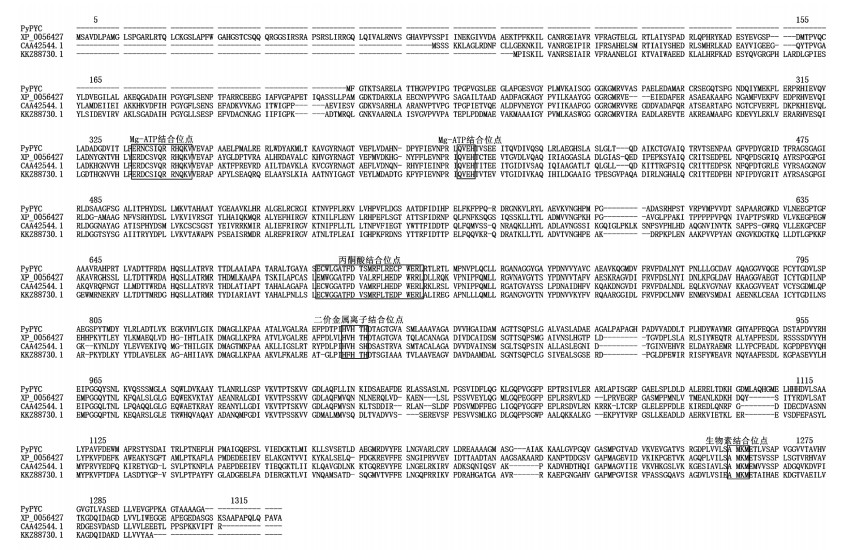 |
| 图 2 来自不同物种的PYC氨基酸序列比对 Fig. 2 Alignment of PYC amino acid sequences from different species 注: 登录号分别为: Coccomyxa subellipsoidea C-169 (XP_005642706.1)、Saccharomyces cerevisiae (CAA42544.1)、Rhizobium phaseoli Ch24-10 (KKZ88730.1) |
在系统进化树上(图 3), PyPYC与来自紫球藻(Porphyridium purpureum)PYC以100%的bootstrap值聚在一起, 然后该分支又以较高的bootstrap值(72%)与来自丝囊霉菌(Aphanomyces invadans)和假微型海链藻(Thalassiosira pseudonana CCMP1335)的PYC组成一个分支。此外, 在进化关系上, 该分支又和来自太平洋邻囊菌(Plesiocystis pacifica)和细菌(Sandaracinaceae bacterium)的PYC聚为一支。这些结果似乎表明, PyPYC与细菌的PYC有较近的亲缘关系。
 |
| 图 3 由PYC氨基酸序列构建的系统发育树 Fig. 3 Phylogenetic tree constructed with the PYC amino acid sequences 注: 登录号分别为: Porphyridium purpureum (KAA8491876.1)、Aphanomyces invadans (XP 008865358.1)、Thalassiosira pseudonana CCMP1335 (EED88205.1)、Plesiocystis pacifica (WP 045116520.1)、Sandaracinaceae bacterium (RZO64306.1)、Chloropicon primus (QDZ22274.1)、Micromonas commoda (XP_002503347.1)、Tetrabaena socialis (PNH00295.1)、Coccomyxa subellipsoidea C-169 (XP_ 005642706.1)、Micractinium conductrix (PSC70059.1)、Saccharomyces cerevisiae S288C (NP 011453.1)、Agrobacterium tumefaciens (WP 003505139.1)、Corynebacterium glutamicum (WP 011013816.1) |
为探究PyPYC基因对不同碳源的响应情况, 我们采用实时荧光定量PCR检测了PyPYC在不同处理中的表达量变化(图 4)。结果显示, 在以NaHCO3作碳源时, PyPYC基因的表达量随NaHCO3浓度升高出现上调趋势, 但和正常浓度碳(2 mmol/L NaHCO3)相比, 差异不显著(图 4a); 而在以CO2为碳源加富处理时, PyPYC基因表达量随着CO2浓度的升高显著升高, 特别是暴露在体积分数为2×10-3 CO2时, 其表达量比暴露在空气高14倍(图 4b)。
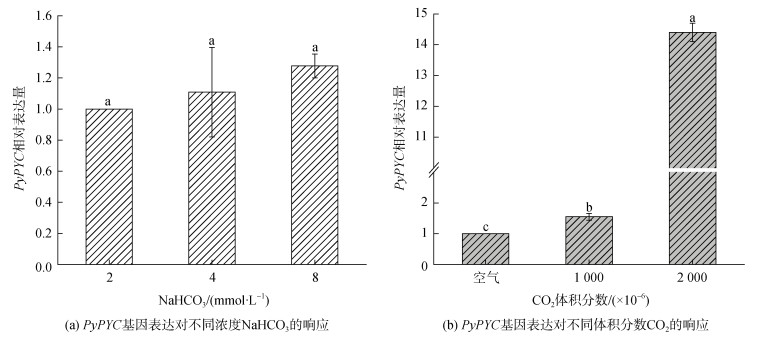 |
| 图 4 PyPYC基因表达对不同碳源的响应 Fig. 4 Relative expression of the PyPYC gene in response to different inorganic carbon sources |
如图 2所示, 来自条斑紫菜的PYC蛋白序列活性位点与来自细菌(Saccharomyces cerevisiae, Rhizobium phaseoli)和微藻(Coccomyxa subellipsoidea C-169)PYC的对应位点序列几乎完全相同, 该结果说明PYC氨基酸序列功能性位点区域在进化上非常保守。
如前所述, 条斑紫菜叶状体在生长过程中由于生境的变化而经历不同类型碳源(暴露在空气中接触空气CO2及没入海水接触海水无机碳)和含水量的变化。前期研究表明, 紫菜叶状体在含水量不低于60%时, 增加空气中CO2能提高紫菜100% CO2固定率[4, 32-33]。因此, 在获得条斑紫菜PYC基因全长序列后, 在保证其含水量不受胁迫(含水量 > 60%)的前提条件下(将紫菜叶状体置于湿润的滤纸上, 见方法部分), 我们模拟叶状体在野外生境的碳源情况, 探究紫菜叶状体中羧化酶PYC基因对不同碳源的响应。结果表明, 相对于碳酸氢根作为主要碳源, 叶状体中的PyPYC基因对CO2的变化更敏感(图 4)。自20世纪70年代以来, 越来越多的证据表明, 大多数藻类存在CO2浓缩机制(CO2 concentrating mechanism, CCM), 该机制协助藻类高效利用海水和空气中的无机碳源[32, 34-38]。而且, 有些海藻的CCM机制中不仅存在碳酸酐酶(carbonic anhydrase, CA), 还发现类似高等植物高效固碳途径-C4途径的一些酶, 如烯醇式丙酮酸羧化酶(Phosphoenolpyruvate Carboxylase, PEPC)和磷酸烯醇式丙酮酸羧激酶(phosphoenolpyruvate carboxykinase, PEPCK)等, 某些特殊生境或培养条件会诱导类C4途径酶发挥固碳作用[39-42]。如在大型绿藻浒苔(Ulva prolifera)中发现, 低光和低CO2水平时主要通过CA利用海水中的HCO3−, 而高光(1 200~2 000 μmol photons·m-2·s-1)时, 则通过PEPC和PEPCK酶利用HCO3−, 从而揭示了浒苔高效固碳机制[43]。
与浒苔不同的是, 条斑紫菜叶状体中PEPC酶活较低, 并且在加富HCO3− (4 mmol/L)人工海水培养时, PEPCK和PYC转录水平表达出现升高趋势, 同时也伴随着PYC羧化酶活和PEPCK脱羧酶活的升高[37]。Shao等[44]研究同样表明, 高碳(0.1 mol/L KHCO3)促进海带(Saccharina japonica)PEPCK脱羧酶活性增加, 但未检测到PEPC酶活。以上结果表明, 大型红藻和褐藻中可能存在与大型绿藻不同的类C4途径, 绿藻以PEPC和PEPCK途径为主, 而红藻和褐藻以PYC和PEPCK为主。莱茵衣藻(Chlamydomonas reinhardtii)在低碳条件培养(培养基通入空气, 即含体积分数为3.60×10-4 CO2), 增加培养基中NH4+后, PYC酶活比PEPC酶活升高, 作者认为该结果证实了以前的研究结论, 即植物细胞内不同的羧化酶提供了OAA合成支路, 为三羧酸循环提供C4复合物, 从而为氮同化和氨基酸合成提供碳骨架[45]。
综上所述, 我们初步认为, 适当高浓度无机碳环境(如增加空气中CO2和增加水体中HCO3−), 能诱导条斑紫菜PYC和PEPCK表达进而协助紫菜进行高效固碳, 而且高浓度CO2更易于诱导PYC的表达。PyPYC基因对高浓度CO2的响应将有助于条斑紫菜叶状体暴露在空气中时高效利用大气中CO2, 至于PyPYC基因对CO2和HCO3-表现出不同的响应则需要更深入的研究。
4 总结来自不同物种(细菌, 古细菌, 微藻及大型海藻) PYC在活性位点区的氨基酸序列比较保守, 而且来自条斑紫菜、紫球藻和假微型海链藻的PYC与细菌的PYC有比较近的亲缘关系。该基因在RNA水平上对环境中CO2的变化响应比对HCO3-更敏感, 推测其在协助条斑紫菜高效利用空气中CO2发挥作用。
| [1] |
YAMAZAKI A, NAKANISHI K, SAGA N. Axenic tissue culture and morphogenesis in Porphyra yezoensis (Bangiales, Rhodophyta)[J]. Journal of Phycology, 1998, 34(6): 1082-1087. DOI:10.1046/j.1529-8817.1998.341082.x |
| [2] |
SAHOO D, TANG X R, YARISH C. Porphyra-the economic seaweed as a new experimental system[J]. Current Science, 2002, 83(11): 1313-1316. |
| [3] |
杨宇峰, 罗洪添, 王庆, 等. 大型海藻规模栽培是增加海洋碳汇和解决近海环境问题的有效途径[J]. 中国科学院院刊, 2021, 36(3): 259-269. YANG Yufeng, LUO Hongtian, WANG Qing, et al. Large-scale cultivation of seaweed is effective approach to increase marine carbon sequestration and solve coastal environmental problems[J]. Bulletin of Chinese Academy of Sciences, 2021, 36(3): 259-269. DOI:10.16418/j.issn.1000-3045.20210217103 |
| [4] |
ZHOU W, HE L W, YANG F, et al. Pyropia yezoensis can utilize CO2 in the air during moderate dehydration[J]. Chinese Journal of Oceanology and Limnology, 2014, 32(2): 358-364. DOI:10.1007/s00343-014-3093-7 |
| [5] |
KITADE Y, TAGUCHI G, SHIN J A, et al. Porphyra monospore system (Bangiales, Rhodophyta): A model for the developmental biology of marine plants[J]. Phycological Research, 1998, 46(1): 17-20. DOI:10.1111/j.1440-1835.1998.tb00092.x |
| [6] |
SAGA N, KITADE Y. Porphyra: A model plant in marine sciences[J]. Fisheries Science, 2002, 68(2): 1075. |
| [7] |
WANG D M, YU X Z, XU K P, et al. Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment[J]. Nature Communications, 2020, 11: 4028. DOI:10.1038/s41467-020-17689-1 |
| [8] |
UTTER M F, KEECH D B. Formation of oxaloacetate from Pyruvate and CO2[J]. Biological Chemistry, 1960, 235: 17-18. DOI:10.1016/S0021-9258(18)69442-6 |
| [9] |
王镜岩, 朱圣庚, 徐长法, 等. 生物化学(第三版下册)[M]. 北京: 高等教育出版社, 2002: 154-158. WANG Jingyan, ZHU Shenggeng, XU Changfa, et al. Biochemistry (Third Edition, Second Volume)[M]. Beijing: Higher Education Press, 2002: 154-158. |
| [10] |
MARIN-VALENCIA I, ROE C R, PASCUAL J M. Pyruvate carboxylase deficiency: mechanisms, mimics and anaplerosis[J]. Molecular Genetics and Metabolism, 2010, 101(1): 9-17. DOI:10.1016/j.ymgme.2010.05.004 |
| [11] |
TOBIAS F, MATTHIAS P, JVRGEN T, et al. Metabolic fluxes in the central carbon metabolism of Dinoroseobacter shibae and Phaeobacter gallaeciensis, two members of the marine Roseobacter clade[J]. BMC Microbiology, 2009, 9(1): 209. |
| [12] |
STUCKA R, DEQUIN S, SALMON J M, et al. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains[J]. MGG Molecular & General Genetics, 1991, 229(2): 307-315. |
| [13] |
ZELLE R M, TRUEHEART J, HARRISON J C et al. Phosphoenolpyruvate carboxykinase as the sole anaplerotic enzyme in Saccharomyces cerevisiae[J]. Applied and Environmental Microbiology, 2010, 76(16): 5383-5389. DOI:10.1128/AEM.01077-10 |
| [14] |
WURTELE E S, NIKOLAU B J. Plants contain multiple biotin enzymes: Discovery of 3-methylcrotonyl-CoA carboxylase, propionyl-CoA carboxylase and pyruvate carboxylase in the plant kingdom[J]. Archives of Biochemistry and Biophysics, 1990, 278(1): 179-186. DOI:10.1016/0003-9861(90)90246-U |
| [15] |
TSUJI Y, SUZUKI I, SHIRAIWA Y. Photosynthetic carbon assimilation in the Coccolithophorid Emiliania huxleyi (Haptophyta): Evidence for the predominant operation of the C3 cycle and the contribution of β - Carboxylases to the active anaplerotic reaction[J]. Plant & Cell Physiology, 2009, 50(2): 318-329. |
| [16] |
PENG H F, WEI D, Chen G, et al. Transcriptome analysis reveals global regulation in response to CO2 supplementation in oleaginous microalga Coccomyxa subellipsoidea C-169[J]. Biotechnology for Biofuels, 2016, 9(1): 151. DOI:10.1186/s13068-016-0571-5 |
| [17] |
TSUJI Y, SUZUKI I, SHIRAIWA Y. Enzymological evidence for the function of a plastid-located pyruvate carboxylase in the Haptophyte alga Emiliania huxleyi: a novel pathway for the production of C4 compounds[J]. Plant & Cell Physiology, 2012, 53(6): 1043-1052. |
| [18] |
ROLEDA M Y and HURD C L. Seaweed responses to ocean acidification[J]. Seaweed Biology, 2012, 219: 407-431. |
| [19] |
XU K, CHEN H Z, WANG W L, et al. Responses of photosynthesis and CO2 concentrating mechanisms of marine crop Pyropia haitanensis thalli to large pH variations at different time scales[J]. Algal Research, 2017, 28: 200-210. DOI:10.1016/j.algal.2017.10.023 |
| [20] |
WANG Y Y, XU K, WANG W L, et al. Physiological differences in photosynthetic inorganic carbon utilization between gametophytes and sporophytes of the economically important red algae Pyropia haitanensis[J]. Algal Research, 2019, 39: 101436. DOI:10.1016/j.algal.2019.101436 |
| [21] |
ZHANG B Y, YANG F, WANG GC, et al. Cloning and quantitative analysis of the carbonic anhydrase gene from Porphyra yezoensis[J]. Journal of Phycology, 2010, 46(2): 290-296. DOI:10.1111/j.1529-8817.2009.00801.x |
| [22] |
HOFMANN K, TMBASE S W. TMBASE-A database of membrane spanning protein segments[J]. Biology Chemistry Hoppe Seyler, 1993, 322(1): 65-68. |
| [23] |
LARKIN M A, BLACKSHIELDS G, BROWN N P, et al. Clustal W and Clustal X version 2.0[J]. Bioinformatics, 2007, 23(21): 2947-2948. DOI:10.1093/bioinformatics/btm404 |
| [24] |
KUMAR S, STECHER G, LI M, et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547-1549. DOI:10.1093/molbev/msy096 |
| [25] |
WU X J, HUANG A Y, WANG C, et al. Variation of expression levels of seven housekeeping genes at different life-history stages in Porphyra yezoensis[J]. PLoS One, 2013, 8(4): 360740. |
| [26] |
ATTWOOD P V. The Structure and the mechanism of action of pyruvate carboxylase[J]. International Journal of Biochemistry and Cell Biology, 1995, 27(3): 231-249. DOI:10.1016/1357-2725(94)00087-R |
| [27] |
JITRAPAKDEE S, WALLACE J C. Structure, function and regulation of pyruvate carboxylase[J]. The Biochemical Journal, 1999, 340(1): 1-16. DOI:10.1042/bj3400001 |
| [28] |
BRANSON J P, NEZIC M, WALLACE J C, et al. Kinetic characterization of yeast pyruvate carboxylase isozyme pyc1[J]. Biochemistry, 2002, 41(13): 4459-4466. DOI:10.1021/bi011888m |
| [29] |
XIANG S, TONG L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction[J]. Nature Structural & Molecular Biology, 2008, 15(3): 295-302. |
| [30] |
VALLE M. Pyruvate carboxylase, structure and function[M]// HARRIS J R, MARLES-WRIGHT J (eds). Macromolecular protein complexes, Subcellular Biochemistry 83. Springer International Publishing Switzerland, 2017, 83(9): 291-322.
|
| [31] |
JITRAPAKDEE S, NEZIC M G, CASSADY A I, et al. Molecular cloning and domain structure of chicken pyruvate carboxylase[J]. Biochemical and Biophysical Research Communications, 2002, 295(2): 387-393. DOI:10.1016/S0006-291X(02)00651-4 |
| [32] |
XU J T, GAO K S. Photosynthetic performance of the red alga Pyropia haitanensis during emersion, with special reference to effects of solar UV radiation, dehydration and elevated CO2 concentration[J]. Photochemistry and Photobiology, 2015, 91(6): 1376-1381. DOI:10.1111/php.12531 |
| [33] |
HUAN L, WANG C, GAO S, et al. Preliminary comparison of atmospheric CO2 enhancement to photosynthesis of Pyropia yezoensis (Bangiales, Rhodophyta) leafy thalli and filamentous thalli[J]. Phycological Research, 2018, 66(2): 117-126. DOI:10.1111/pre.12213 |
| [34] |
GAO K S, ARUGA Y, ASADA K, et al. Enhanced growth of the red alga Porphyra yezoensis Ueda in high CO2 concentrations[J]. Journal of Applied Phycology, 1991, 3(4): 355-362. DOI:10.1007/BF02392889 |
| [35] |
岳国峰, 周百成. 条斑紫菜对无机碳的利用[J]. 海洋与湖沼, 2000, 31(3): 246-251. YUE Guofeng, ZHOU Baicheng. Inorganic carbon utilization by Pyropia yezoensis Ueda[J]. Oceanologia et Limnologia Sinica, 2000, 31(3): 246-251. DOI:10.3321/j.issn:0029-814X.2000.03.003 |
| [36] |
BAO M L, WANG J H, XU T p, et al. Rising CO2 levels alter the responses of the red macroalga Pyropia yezoensis under light stress[J]. Aquaculture, 2019, 501: 325-330. DOI:10.1016/j.aquaculture.2018.11.011 |
| [37] |
ZHANG B Y, XIE X J, LIU X H, et al. The carbonate concentration mechanism of Pyropia yezoensis (Rhodophyta): evidence from transcriptomics and biochemical data[J]. BMC Plant Biology, 2020, 20(1): 424. DOI:10.1186/s12870-020-02629-4 |
| [38] |
ZHANG D, XU J T, BAO M L, et al. Elevated CO2 concentration alleviates UVR-induced inhibition of photosynthetic light reactions and growth in an intertidal red macroalga[J]. Journal of Photochemistry & Photobiology, B: Biology, 2020, 213: 112074. |
| [39] |
ROBERTS K, GRANUM E, LEEGOOD R C, et al. C3 and C4 pathways of photosynthetic carbon assimilation in marine diatoms are under genetic, not environmental, control[J]. Plant Physiology, 2007, 145(1): 230-235. DOI:10.1104/pp.107.102616 |
| [40] |
MORONEY J V, MA Y B, FREY W D, et al. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles[J]. Photosynthesis Research, 2011, 109(3): 133-149. |
| [41] |
LI W, HAJJAMI M E, CHEN S, et al. Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica[J]. Biotechnology for Biofuels, 2019, 12(1): 1-21. DOI:10.1186/s13068-018-1346-y |
| [42] |
ZHANG H, ZHOU Y P, LIU T Q, et al. Initiation of efficient C4 pathway in response to low ambient CO2 during the bloom period of a marine dinoflagellate[J]. Environmental Microbiology, 2021, 23(6): 3196-3211. DOI:10.1111/1462-2920.15545 |
| [43] |
LIU D Y, MA Q, IVAN V, et al. Role of C4 carbon fixation in Ulva prolifera, the macroalga responsible for the world's largest green tides[J]. Communications Biology, 2020, 3(1): 494. DOI:10.1038/s42003-020-01225-4 |
| [44] |
SHAO Z R, WANG W L, ZHANG P Y, et al. Genome-wide identification of genes involved in carbon fixation in Saccharina japonica and responses of putative C4-related genes to bicarbonate concentration and light intensity[J]. Plant Physiology and Biochemistry, 2019, 137: 75-83. DOI:10.1016/j.plaphy.2019.01.032 |
| [45] |
GIORDANO M, NORICI A, FORSSEN M, et al. An anaplerotic role for mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii[J]. Plant Physiology, 2003, 132: 2126-2134. DOI:10.1104/pp.103.023424 |
 2022, Vol. 46
2022, Vol. 46