
涓浗娴锋磱婀栨布瀛︿細涓诲姙銆�
鏂囩珷淇℃伅
- 寰愬湥閽�, 鏋楀媺, 闂澗鏉�, 鍙查洦绾�, 鑻椾寒, 鏉庢槑浜�, 闄堢偗. 2016.
- XU Sheng-Zhao, LIN Mian, YAN Song-Song, SHI Yu-Hong, Miao Liang, LI Ming-Yun, CHEN Jiong. 2016.
- 澶ч粍楸�(Larimichthys crocea)鏂板搧绉嶁€滀笢娴�1鍙封€濅綋闀跨浉鍏崇殑DArT鏍囪绛涢€�
- IDENTIFICATION OF BODY-LENGTH-RELATED DART MARKERS IN NEW CALTIVAR "DONGHAI NO.1" OF LARGE YELLOW CROAKER LARIMICHTHYS CROCEA
- 娴锋磱涓庢箹娌�, 47(5): 1047-1054
- Oceanologia et Limnologia Sinica, 47(5): 1047-1054.
- http://dx.doi.org/10.11693/hyhz20160600128
-
鏂囩珷鍘嗗彶
- 鏀剁ǹ鏃ユ湡锛�2016-06-20
- 鏀朵慨鏀圭ǹ鏃ユ湡锛�2016-07-11
澶ч粍楸�(Larimichthys crocea)灞炵‖楠ㄩ奔绾层€侀矆褰㈢洰銆佺煶棣栭奔绉戙€侀粍楸煎睘, 鏄垜鍥解€滃洓澶ф捣姘寸粡娴庨奔绫烩€濅箣涓€(濮氬悍绛�, 2008)銆�20涓栫邯90骞翠唬浜哄伐绻佽偛鎴愬姛鍚�, 澶ч粍楸兼垚涓烘垜鍥戒汉宸ヨ偛鑻楅噺鍜屽吇娈栬妯℃渶澶х殑娴锋磱姘翠骇鍏绘畺鍝佺, 鍏剁粡娴庢晥鐩婃樉钁椼€傜敱浜庝笉閲嶈绉嶈川淇濇姢鍜屼汉宸ラ€夎偛, 鐩墠浜哄伐鍏绘畺澶ч粍楸煎湪浣撳舰銆佺敓闀裤€佽倝璐ㄣ€佹€ф垚鐔熴€佹姉閫嗘€с€佹姉鐥呮€х瓑澶氶挓鎬х姸涓婂嚭鐜颁簡琛伴€€, 閬椾紶澶氭牱鎬ч檷浣�, 鍦ㄤ竴瀹氱▼搴︿笂闄愬埗浜嗗叾鍏绘畺涓氱殑鍙寔缁彂灞�(Gao et al, 2010)銆傚洜姝�, 鏈夊繀瑕佸澶ч粍楸艰繘琛屽搧璐ㄦ敼鑹�, 鍩硅偛鍑哄叿鏈夌敓闀垮揩銆佹姉鐥呭己銆佷綋鍨嬭倝璐ㄥソ銆佽€愪綆娓╃瓑鎬х姸鐨勪紭鑹搧绉嶃€�
DNA鍒嗗瓙鏍囪鏄牴鎹釜浣撻棿鍩哄洜缁凞NA鐨勫鎬佹€у彂灞曡捣鏉ョ殑涓€绫婚仐浼犳爣璁版妧鏈�, 鐩告瘮褰㈡€佸鏍囪銆佺粏鑳炲鏍囪銆佺敓鐗╁寲瀛︽爣璁板叿鏈変竴瀹氱殑浼樿秺鎬�(榄忎笢鏃虹瓑, 2001), 宸插箍娉涘簲鐢ㄤ簬姘翠骇鍔ㄧ墿閬椾紶鑲茬銆傚ぇ榛勯奔閬椾紶鑲茬涔熸櫘閬嶉噰鐢―NA鍒嗗瓙鏍囪鎶€鏈€�鐜嬪織鍕囩瓑(2002)閲囩敤AFLP鎶€鏈�(Amplified fragment length polymorphism)鍒嗘瀽鏄剧ず, 澶ч粍楸奸噹鐢熺兢浣撳拰2涓吇娈栫兢浣�, 2涓寤哄吇娈栧ぇ榛勯奔缇や綋鐨勯仐浼犲鏍锋€т綆浜庨噹鐢熺兢浣撱€�鏉庢槑浜戠瓑(2003)閲囩敤RAPD鎶€鏈�(Random amplified polymorphic DNA)鍒嗘瀽琛ㄦ槑, 璞″北娓綉绠卞吇娈栧ぇ榛勯奔缇や綋閬椾紶澶氭牱鎬ф按骞宠緝浣庛€�鏉庨箯椋炵瓑(2008)閲囩敤楸肩嚎绮掍綋DNA鐨勭粏鑳炶壊绱�b(Cytochrome b, Cytb)鍩哄洜鐗囨澶氭€佹€у彲浠ュ尯鍒嗗ぇ榛勯奔銆侀驳楸煎拰缇庡浗绾㈤奔銆�鐜嬫檽娓呯瓑(2008)閲囩敤AFLP鍜孲SR鎶€鏈�(Simple sequence repeats)瀵逛翰鏈笌鏉備氦瀛愪唬鍒嗘瀽缁撴灉琛ㄦ槑, 鏉備氦瀛愪唬涓庢瘝鏈ぇ榛勯奔涔嬮棿鐨勯仐浼犲悓璐ㄦ€ф瀬楂�, 灞炰簬寮傛簮绮惧瓙璇卞澶ч粍楸奸泴鏍稿彂鑲蹭釜浣撱€�瀹佸渤(2007)鍒嗙鑾峰緱鐨凙FLP鍜孲SR鏍囪搴旂敤浜庡ぇ榛勯奔鐨勯泴鎬у拰闆勬€ц繛閿佸浘璋辩殑鏋勫缓, 鍚屾椂纭畾浜嗗ぇ榛勯奔鎬у埆鍐冲畾鏈哄埗銆傛澶�, 涔熼檰缁瓫閫夎幏寰楄澶氭湁浠峰€肩殑SSR鏍囪(Guo et al, 2005; Chang et al, 2009)銆傛棭鏈烡NA鍒嗗瓙鏍囪鎶€鏈富瑕佸簲鐢ㄤ簬澶ч粍楸奸仐浼犲鏍锋€ф娴嬨€佺郴璋辩‘璁ゃ€侀仐浼犺繛閿佸浘璋辩殑鏋勫缓涓�(Ye et al, 2014)銆傝繎骞存潵, 鐮旂┒鑰呬富瑕佽嚧鍔涗簬閲囩敤DNA鍒嗗瓙鏍囪鎶€鏈瓫閫夋€х姸鐩稿叧鏍囪銆傚垬璐ゅ痉绛夐噰鐢ㄥ井鍗槦鏍囪鎶€鏈涓嶅悓澶ч粍楸煎绯诲拰缇や綋杩涜鍒嗘瀽, 绛涢€夊埌涓庡ぇ榛勯奔鐢熼暱鎬х姸绱у瘑鐩稿叧鐨勫井鍗槦鏍囪(鍒樿搐寰风瓑, 2012, 2013; 鍙跺崕绛�, 2014)銆傝枦鑹箟绛�(2013)鐮旂┒琛ㄦ槑澶ч粍楸艰倢鑲夌敓闀挎姂鍒剁礌鍩哄洜3鈥欑闈炵紪鐮佸尯寰崼鏄熷簭鍒楀鎬佹€т笌澶ч粍楸间綋闀裤€佷綋璐ㄩ噺涔嬮棿鐨勭浉鍏崇郴鏁版病鏈夎揪鍒版樉钁楁按骞炽€備絾鐢熼暱鎬х姸鐩稿叧鏍囪鍙戞帢杈冨皯, 姝ゅ鏄惁鍙敤浜庣敓浜у疄璺佃繕闇€杩涗竴姝ラ獙璇�, 鍥犳杩橀渶缁х画绛涢€夌敓闀挎€х姸鐩稿叧鏍囪銆�
澶氭牱鎬ц姱鐗囨妧鏈�(Diversity arrays technology, DArT)鏄竴绉嶅熀浜庡熀鍥犺姱鐗囨妧鏈殑DNA鎸囩汗鍥捐氨鍒嗘瀽鏂规硶, 鍙箍娉涚敤浜庢娴嬪拰鍒嗘瀽鍔ㄧ墿銆佹鐗╁拰寰敓鐗╃殑DNA宸紓浠ュ強鏋勫缓閬椾紶鍥捐氨銆丵TL瀹氫綅鍜屽搧绉嶆寚绾瑰浘璋遍壌瀹�(S谩nchez-Sevilla et al, 2015)銆備笌甯歌鎶€鏈浉姣�, DArT涓嶉渶瑕佹槑纭墿绉嶇殑鍩哄洜缁凞NA搴忓垪淇℃伅, 鍏锋湁楂橀€氶噺鍜屼綆鎴愭湰鐨勬樉钁楃壒鐐�, 鍏嬫湇浜嗕互寰€璺戠數娉冲嚌鑳朵负涓荤殑鏍囪鎶€鏈骇閲忎綆銆佹垚鏈珮銆佽€楁椂闀裤€佽嚜鍔ㄥ寲绋嬪害浣庣瓑缂虹偣, 鍙敤灏戦噺鎴愭湰灏卞彲杩涜鍏ㄥ熀鍥犵粍鐨勯珮閫氶噺鍥捐氨鍒嗘瀽銆傜洰鍓嶈鎶€鏈凡鎴愬姛鐢ㄤ簬姘寸ɑ銆佸ぇ楹︺€佸皬楹︺€佹补鑿溿€佹鏍戙€佽嫻鏋溿€佹湪钖€佹嫙鍗楄姤銆佹湪璞嗐€佸ぇ楹︾梾鍘熻弻銆佹矙闂ㄦ皬鑿岀瓑鐢熺墿鐨勯仐浼犺繛閿佸浘璋变互鍙婂熀鍥犲畾浣嶇爺绌朵腑(Hackl et al, 2010; Schouten et al, 2012)銆傚湪姘翠骇鍔ㄧ墿鐨勭爺绌朵腑浠呰璇ユ妧鏈簲鐢ㄤ簬涓夌枺姊瓙锜瑰湴鐞嗙缇ゅ鏍锋€у垎鏋�(鑽f檾濠х瓑, 2014)銆�
鏈爺绌舵棬鍦ㄩ噰鐢―ArT鎶€鏈壌瀹氫笌澶ч粍楸间綋闀跨浉鍏崇殑DArT鏍囪銆傞鍏堟寜鍒嗙缇や綋鏍囪鍏宠仈鍒嗘瀽娉曠瓫閫夆€滀笢娴�1鍙封€濆ぇ榛勯奔浣撻暱鐩稿叧鐨凞ArT鏍囪, 鍚庣画杩涗竴姝ラ獙璇佸叾鐩稿叧鎬�, 浠ユ湡涓哄ぇ榛勯奔閫夎偛鍜岀璐ㄨ祫婧愬埄鐢ㄦ彁渚涙湁鐢ㄥ弬鑰冭祫鏂欍€�
1 鏉愭枡涓庢柟娉� 1.1 鏍峰搧閲囬泦2012骞�11鏈堜粠瀹佹尝璞″北娓咕姘翠骇鑻楃鏈夐檺鍏徃缃戠鍏绘畺鐨勨€滀笢娴�1鍙封€濆ぇ榛勯奔(1榫�)涓寫閫夊仴搴锋棤鎹熶激鐨勫ぇ榛勯奔199灏�, 娴嬮噺姣忔潯楸肩殑浣撻暱, 骞跺湪楸奸硟鐩栧唴渚ц緝杞儴浣嶆鍏ョ數瀛愭爣璁般€傜劧鍚庡垎鍒皢鍏舵斁鍏ユ按娉ユ睜涓殏鍏汇€備娇鐢⊿PSS 17.0杞欢缁熻鍒嗘瀽, 鍒嗗埆寤虹珛浣撻暱鐨勬鎬佸垎甯冨浘, 鍙栦綋闀夸綅浜�10%鐨勯珮鍊间釜浣撹涓衡€滄瀬绔ぇ缇や綋鈥�(20灏�), 10%浣庡€间釜浣撹涓衡€滄瀬绔皬缇や綋鈥� (20灏�)銆�
1.2 鍩哄洜缁勪唬琛ㄦ€NA鐗囨鏂囧簱鏋勫缓鏂囧簱鏋勫缓鏂规硶鍙傜収Jaccoud绛�(2001)鎻忚堪銆傞噰鐢ㄩ厷-姘豢娉曟彁鍙栧熀鍥犵粍DNA, 骞跺皢鈥滄瀬绔ぇ缇や綋鈥濆拰鈥滄瀬绔皬缇や綋鈥濈殑澶ч粍楸煎熀鍥犵粍DNA绛夐噺娣峰悎銆�500ng娣峰悎鍩哄洜缁凞NA鐢ㄥ垏鍓查鐜囦綆鐨勯檺鍒舵€у唴鍒囬叾Pst鈪犲垎鍒笌鍒囧壊棰戠巼楂樼殑Alu鈪犮€�Ban鈪°€�Bsp1286I銆�BstNI銆�Hae鈪€€�Rsa鈪犲拰Taq鈪犵粍鍚堣繘琛岄叾鍒囥€傚湪T4 DNA杩炴帴閰朵綔鐢ㄤ笅, 绾寲鐨凞NA杩炴帴涓�Pst鈪犵壒寮傛€ф帴澶�(Wenzl et al, 2004)銆傝繛鎺ヤ骇鐗╀綔涓烘ā鏉跨敤浜庡悗缁璓CR鎵╁, 鎵€鐢ㄥ紩鐗╀负DArT-Pst鈪犲紩鐗�(Wenzl et al, 2004), 鍙嶅簲绋嬪簭濡備笅: 94掳C鍙樻€�5min鍚�, 浠ヤ笅绋嬪簭閲嶅35涓惊鐜�, 94掳C鍙樻€�30s, 53掳C澶嶆€�30s, 72掳C寤朵几1min, 寰幆瀹屾垚鍚�72掳C寤朵几鍙嶅簲10min銆傛墿澧炰骇鐗╁厠闅嗚嚦杞戒綋pMD19-T, 杞寲澶ц偁鏉嗚弻TOP10F骞舵秱甯冧簬鍚皑鑻勯潚闇夌礌鍜孹-gal鐨凩B鍩瑰吇鍩轰笂銆�
1.3 鍩哄洜缁凞NA澶嶆潅鎬ч檷浣庢柟娉曠殑浼樺寲 1.3.1 鎺㈤拡鍒跺鍙婅姱鐗囩偣鍒�浠庝唬琛ㄦ€у熀鍥犵粍DNA鏂囧簱涓殢鏈烘寫閫夊崟鑿岃惤, 鍒╃敤璐ㄧ矑杞戒綋涓婄殑閫氱敤寮曠墿M13F-47鍜孧13R-48瀵规彃鍏ョ殑DNA鐗囨杩涜PCR鎵╁, 浜х墿鐢�1鍊嶄綋绉紓涓欓唶娌夋穩銆傛瘡涓�7绉嶉檷浣庡熀鍥犵粍澶嶆潅鎬ф柟娉曞熀鍥犵粍浠h〃鎬NA鐗囨鏁颁负840涓€傚湪鑺墖涓竷缃川鎺ф帰閽�, 闃存€у鐓ф帰閽�, 闃虫€у鐓ф帰閽�, 绌虹櫧瀵圭収鎺㈤拡, 宸ヤ綔鎺㈤拡銆傛瘡涓帰閽堝寘鍚笁涓噸澶嶃€傛瘡寮犺姱鐗囧寘鍚�4涓偣闃�, 姣忎釜鐐归樀25琛屻€�27鍒�, 姣忎釜鐐圭殑浣嶇疆鐢ㄢ€滆鏍�-鍒楁爣鈥濊〃绀恒€傚叾涓�1-1鈥�1-3涓鸿川鎺ф帰閽�, 1-19鈥�1-21涓洪槼鎬у鐓ф帰閽�, 1-7鈥�1-18銆�1-22鈥�2-3涓洪槾鎬у鐓ф帰閽�, 2-4鈥�25-12涓哄伐浣滄帰閽�, 1-4鈥�1-6銆�25-13鈥�25-27涓虹┖鐧藉鐓ф帰閽堛€備粠Pst鈪�/Alu鈪犮€�Pst鈪�/Ban鈪°€�Pst鈪�/Bsp1286I銆�Pst鈪�/BstNI銆�Pst鈪�/Hae鈪€€�Pst鈪�/Rsa鈪犮€�Pst鈪�/Taq鈪犳枃搴撲腑鑾峰緱鐨�840涓熀鍥犵粍浠h〃鎬NA鐗囨渚濇鎺掑竷鍦�4涓偣闃典腑銆傞噰鐢ㄦ櫠鑺疭martArrayerTM 48鐐规牱浠繘琛岀偣鍒躲€�
1.3.2 鑽у厜鏍囪鍩哄洜缁勪唬琛ㄦ€NA鐗囨鐨勫埗澶�鐢∕13F-47鍜孧13R-48寮曠墿瀵规湭鎻掑叆澶栨簮DNA鐗囨鐨勮浇浣撹繘琛孭CR鎵╁, 璇CR浜х墿浣滀负reference DNA銆傜粡涔欓唶娌夋穩鍚�, 鍔犲叆20渭L鐏弻姘存憾瑙�, 鏀锯€�30掳C鍐扮淇濆瓨澶囩敤銆�
閲囩敤閰�-姘豢娉曟彁鍙栧ぇ榛勯奔鈥滄瀬绔ぇ缇や綋鈥濅笌鈥滄瀬绔皬缇や綋鈥濆熀鍥犵粍DNA, 涓ょ粍DNA鍏堝垎鍒娇鐢�7缁勯檺鍒舵€у唴鍒囬叾(Pst鈪�/Alu鈪犮€�Pst鈪�/Ban鈪°€�Pst鈪�/Bsp1286I銆�Pst鈪�/BstNI銆�Pst鈪�/Hae鈪€€�Pst鈪�/Rsa鈪犮€�Pst鈪�/Taq鈪�)闄嶄綆鍩哄洜缁勫鏉傛€�, 鍐嶅姞Pst鈪犵壒寮傛€ф帴澶�, 鎺ョ潃鐢�Pst鈪犲紩鐗╁鍔犳帴澶翠骇鐗╄繘琛孭CR鎵╁, 鏈€鍚庢墿澧炰骇鐗╀娇鐢―NAmate娌夋穩娴撶缉10鍊嶄綋绉€�
鍙�150ng鍩哄洜缁勪唬琛ㄦ€NA鐗囨鍙樻€у悗鐢―ecaLabel DNA Labeling Kit杩涜Cy3鏍囪, 鍔犲叆鍚崄纰卞熀闅忔満寮曠墿鐨�5脳缂撳啿娑层€丮ixC銆丆y5-dCTP銆乪xo-Klenow fragment鍏�2.1渭L, 37掳C瀛佃偛10min鍚�, 鍔犲叆dNTPs 0.4渭L, 37掳C瀛佃偛30min, 鍔犲叆0.1渭L EDTA (pH 8.0)缁堟鍙嶅簲銆傚彇150ng reference DNA鍙樻€у悗鐢―ecaLabel DNA Labeling Kit杩涜Cy5鏍囪銆�
1.3.3 鑺墖鏉備氦Cy3鍜孋y5鏍囪鍙嶅簲浜х墿娣峰悎, 鍐嶅姞鍏�1渭L椴戠簿DNA(10g/L)鍜�50渭L ExpressHyb鈩㈡潅浜ゆ恫娣峰悎鍚�, 96掳C鍙樻€�3min, 鍐版荡楠ゅ喎1min銆傚皢涓婅堪鍙嶅簲浜х墿鍔犲叆棰勫鐞嗙殑DArT鑺墖涓�, 鍦ㄦ櫠鑺�庐鏉備氦浠腑杩涜鏉備氦(65掳C瀛佃偛杩囧銆傛潅浜ゅ悗鍏堢敤0.3脳SSC, 0.1% SDS娓呮礂涓€娆�, 鍐嶇敤0.06脳SSC娓呮礂涓ゆ, 绂诲績鐢╁共銆�
1.3.4 鑺墖鐨勬壂鎻忎笌鏁版嵁澶勭悊鏉備氦鍚庨噰鐢ㄦ櫠鑺� LuxScanTM 10K-A鍙岄€氶亾婵€鍏夊叡鑱氱劍鎵弿浠繘琛屾壂鎻�, 骞剁敤LuxScan3.0杞欢杩涜鏁版嵁鐨勬彁鍙栥€傝嫢瀵瑰簲鐨剅eference DNA鏉備氦鑽у厜寮哄害杈冨急, 鍒欏睘浜庡潖鐐�, 寮冧箣銆傝川閲忕鍚堟潯浠剁殑鎺㈤拡, 鍏跺搴旂殑鑽у厜寮哄害鎸夌収lg[Cy3 Target/Cy5 Reference]杩涜璁$畻鍧囦竴鍖栥€傚埄鐢ㄦā绯奀-鍧囧€艰仛绫诲垎鏋愭硶(妯$硦搴︿负1.5)灏嗗綊涓€鍖栧悗鐨勮姱鐗囦俊鍙峰€煎垎涓�2缁勮仛绫荤皣(cluster), 濡傛灉璁$畻鍑虹殑鑱氱被绨囦箣闂存柟宸嚦灏戝ぇ浜庢€绘柟宸殑80%, 鍒欒涓烘鎺㈤拡鍏锋湁澶氭€佹€�, 妯$硦C-鍧囧€艰仛绫诲垎鏋愭硶鍙皢鍏跺湪涓嶅悓鑺墖鏍锋湰鍐呭垎鎴�0/1涓ょ粍绫诲埆(Wenzl et al, 2004)銆傞噰鐢ˋNOVA鍗曚晶妫€楠�(one-way ANOVA)鍒嗘瀽缁勫樊寮傛爣璁般€傝绠�P鍊笺€�q鍊煎拰FDR, 骞跺埗浣滅浉搴旂殑鏁g偣鍥俱€�
1.4 澶ч粍楸肩敓闀跨浉鍏冲垎瀛愭爣璁扮殑绛涢€�浠�Pst鈪�/Rsa鈪犱唬琛ㄦ€у熀鍥犵粍DNA鏂囧簱涓殢鏈烘寫閫�3360涓厠闅�, PCR鎵╁鎻掑叆鐗囨, 閲嶆柊鐐瑰埗鑺墖銆傚湪鑺墖涓竷缃川鎺ф帰閽�, 闃存€у鐓ф帰閽�, 闃虫€у鐓ф帰閽�, 绌虹櫧瀵圭収鎺㈤拡, 宸ヤ綔鎺㈤拡銆傛瘡涓帰閽堝寘鍚笁涓噸澶嶃€傛瘡寮犺姱鐗囧寘鍚�16涓偣闃�, 姣忎釜鐐归樀鐨勮缃悓1.3.1銆傚悇鍙�500ng鍩哄洜缁凞NA, 鍒嗗埆鐢�Pst鈪犱笌Rsa鈪犵粍鍚堣繘琛屽弻閰跺垏銆傞叾鍒囧悗鍔�Pst鈪犵壒寮傛€ф帴澶�, 鐢―ArT-Pst鈪犲紩鐗╁紩鐗╁閰跺垏浜х墿杩涜PCR鎵╁銆傝崸鍏夋爣璁板熀鍥犵粍DNA浠h〃鎬х墖娈电殑鍒跺鏂规硶鍚�1.3.2銆傝姱鐗囬澶勭悊銆佹潅浜ゃ€佹壂鎻忋€佹暟鎹彁鍙栧強鏁版嵁鍒濇澶勭悊鏂规硶鍚�1.3.3鍜�1.3.4銆�
1.5 澶ч粍楸肩敓闀跨浉鍏�DArT鏍囪鐨勯獙璇�鍦ㄥ悗鏈熼獙璇佸疄楠屼腑, 閲嶆柊鍙�177灏剧綉绠卞吇娈栫殑鍋ュ悍銆佹棤鎹熶激1榫勨€滀笢娴�1鍙封€濆ぇ榛勯奔銆備娇鐢⊿PSS 17.0杞欢缁熻鍒嗘瀽寤虹珛浣撻暱鐨勬鎬佸垎甯冨浘銆傝繖涓兢浣撴瘡涓釜浣撳熀鍥犵粍DNA浠h〃鎬х墖娈靛埗澶囥€佽姱鐗囬澶勭悊銆佹潅浜ゃ€佹壂鎻忋€佹暟鎹彁鍙栧強鏁版嵁鍒濇澶勭悊鏂规硶鍚�1.3.2鈥� 1.3.4銆傜瓫閫夎幏寰楃殑鍊欓€塂ArT鏍囪杩涜娴嬪簭, 鍚屾椂鐢˙LAST2GO (http://www.blast2go.org)杞欢瀵归壌瀹氱殑DArT鏍囪杩涜鍩哄洜鍔熻兘娉ㄩ噴銆�
2 缁撴灉 2.1 澶ч粍楸间綋闀挎暟鎹粺璁�闅忔満閫夋嫨199涓ぇ榛勯奔鏍锋湰娴嬮噺鍏朵綋闀挎暟鎹€傚疄楠岀兢浣撲腑鐨勪綋闀挎渶澶у€间负16.30cm銆佹渶灏忓€间负11.50cm, 鍧囧€间负13.45cm, 鏍囧噯鍋忓樊1.0337銆傜粡杩嘢hapiro-Willie杩囩▼杩涜姝f€佸垎甯冩楠�, 宄板害涓�0.192, 鍋忓害涓衡€�0.210, 璁$畻寰楀埌P=0.257, 鏍峰搧绗﹀悎姝f€佸垎甯�(P > 0.05)銆傚洜鑰屾牴鎹噰鐢ㄥ垎绂荤兢浣撳垎缁勫垎鏋愭硶(Bulked Segregate Analysis, BSA)寤虹珛鍏充簬浣撻暱鐨勬鎬佸垎甯冨浘(鍥� 1), 鍦ㄥ悇缇や綋涓€夊彇10%鐨勯珮鍊间釜浣撳嵆浣撻暱澶т簬14.80 cm鐨勮涓烘瀬绔ぇ缇や綋, 閫夊彇10%鐨勪綆鍊间釜浣撳嵆浣撻暱灏忎簬12.00 cm鐨勮涓烘瀬绔皬缇や綋, 涓ょ粍涔嬮棿浣撻暱宸紓鏋佹樉钁�(P < 0.01)銆傚彲鐢ㄤ簬涓庣敓闀跨浉鍏矰ArT鏍囪鐨勫垵姝ョ瓫閫夈€�
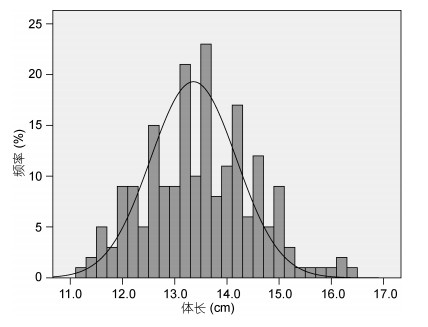 |
| 鍥� 1 澶ч粍楸间綋闀挎鎬佸垎甯冮鐜囩洿鏂瑰浘 Fig. 1 Normal distribution frequency histogram of body length of large yellow croaker |
鏈爺绌朵腑澶ч粍楸煎熀鍥犵粍DNA瀹屾暣, 绾害楂�, 鏃燫NA姹℃煋(鍥� 2A)銆傚皢涓嶅悓浣撻暱鐨勫ぇ榛勯奔娣峰悎鍚�, 鍒嗗埆鐢�Pst鈪�/Alu鈪犮€�Pst鈪�/Ban鈪°€�Pst鈪�/Bsp1286I銆�Pst鈪�/ BstNI銆�Pst鈪�/Hae鈪€€�Pst鈪�/Rsa鈪犮€�Pst鈪�/Taq鈪犻叾鍒囧悗绾寲(鍥� 2B)銆傚湪T4 DNA杩炴帴閰朵綔鐢ㄤ笅, 绾寲鐨凞NA閰跺垏鐗囨涓�Pst 鈪犵壒寮傛€ф帴澶磋繛鎺ャ€傝繛鎺ヤ骇鐗╀綔涓烘ā鏉胯繘琛屽悗缁璓CR鎵╁(鍥� 2C)銆侾CR浜х墿鍏嬮殕鑷硃MD19-T杞戒綋鍚庤浆鍖栧ぇ鑲犳潌鑿孴op10F鈥�, 鎵€寰�7涓熀鍥犵粍浠h〃鎬NA鏂囧簱婊村害鈮�105(琛� 1), 闃虫€у厠闅嗗钩鍧囨彃鍏ョ墖娈甸暱搴� > 500bp(鍥� 2D鈥擩), 绗﹀悎DArT鑺墖鐐瑰埗瑕佹眰銆�
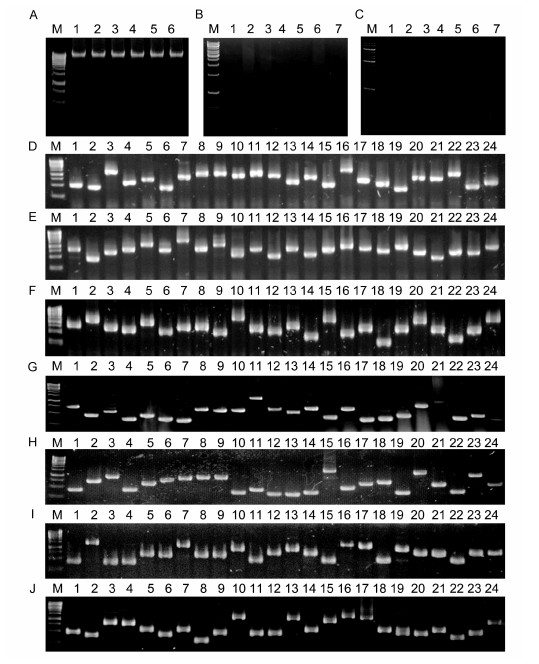 |
| 鍥� 2 澶ч粍楸煎熀鍥犵粍浠h〃鎬NA鐗囨鏂囧簱鏋勫缓 Fig. 2 Construction of genomic representations library of large yellow croaker 娉�: A:涓嶅悓浣撻暱澶ч粍楸煎熀鍥犵粍DNA, M: 1kb DNA ladder, A1: x725, A2: x784, A3: x798, A4: x658, A5: x625; A6: x827; B:娣峰悎鍩哄洜缁凞NA閰跺垏妫€娴�(5渭L), M: 1kb DNA ladder, B1: Pst鈪犲拰Alu鈪�, B2: Pst鈪犲拰Ban鈪�, B3: Pst鈪犲拰Bsp1286I, B4: Pst鈪犲拰BstNI, B5: Pst鈪犲拰Hae鈪�, B6: Pst鈪犲拰Rsa鈪�, B6: Pst I鍜�Taq I; C:鍔�pst I鎺ュご鍚嶱CR鎵╁; D鈥擩: 7涓熀鍥犵粍DNA浠h〃鎬ф枃搴撴彃鍏ョ墖娈电數娉虫娴嬬粨鏋�, 1鈥�24:闅忔満閫夊彇鐨勫厠闅� |
| 鍚嶇О | 搴撳 | 骞冲潎鎻掑叆鐗� 娈靛ぇ灏�(bp) |
澶氭€佹€� 鍏嬮殕鏁� |
澶氭€佹€х巼 (%) |
| Pst鈪�/Alu鈪犳枃搴� | 8.0脳105 | 886 | 58 | 6.90 |
| Pst鈪�/Ban鈪℃枃搴� | 1.0脳106 | 691 | 117 | 13.93 |
| Pst鈪�/Bsp1286I鏂囧簱 | 4.0脳106 | 820 | 68 | 8.10 |
| Pst鈪�/BstNI鏂囧簱 | 1.2脳106 | 607 | 119 | 14.17 |
| Pst鈪�/Hae鈪㈡枃搴� | 4.0脳106 | 623 | 110 | 13.10 |
| Pst鈪�/Rsa鈪犳枃搴� | 4.0脳106 | 765 | 148 | 17.62 |
| Pst鈪�/Taq鈪犳枃搴� | 1.2脳106 | 721 | 140 | 16.67 |
澶ч粍楸尖€滄瀬绔ぇ缇や綋鈥濅笌鈥滄瀬绔皬缇や綋鈥濇牱鍝佹彁鍙栧熀鍥犵粍DNA, 鍒嗗埆閲囩敤7绉嶉叾鍒囩粍鍚�(Pst鈪�/Alu鈪犮€�Pst鈪�/Ban鈪°€�Pst鈪�/Bsp1286I銆�Pst鈪�/BstNI銆�Pst鈪�/Hae鈪€€�Pst鈪�/Rsa鈪犮€�Pst鈪�/Taq鈪�)闄嶄綆鍩哄洜缁勫鏉傛€с€傜粡鏍囪鍚庣殑DNA鐗囨鍒嗗埆涓庡悇鑷狣ArT鑺墖鏉備氦, 鏉備氦缁撴灉娓呮櫚鍙潬銆�7绉嶉叾鍒囩粍鍚堜腑, Pst鈪�/Rsa鈪犱笉浠呭彲闄嶄綆鍩哄洜缁勫鏉傛€�, 涓斿鎬佹€х巼鏈€楂�(17.62%)(琛� 1)銆傚洜姝�, 閫夋嫨Pst鈪�/Rsa鈪犲熀鍥犵粍浠h〃鎬NA鐗囨鏂囧簱鐢ㄤ簬澶ч粍楸间綋闀跨浉鍏矰ArT鏍囪鐨勭瓫閫夈€�
2.4 澶ч粍楸肩敓闀跨浉鍏矰ArT鏍囪鐨勭瓫閫�閲嶆柊鐐瑰埗鐨�Pst鈪�/Rsa鈪� DArT鑺墖, 鍏跺厠闅嗘暟澧炲姞鑷�3360涓�(鍥� 3A)銆傚苟鎸夌収涓婅堪鏂规硶鍒跺鈥滄瀬绔ぇ缇や綋鈥濅笌鈥滄瀬绔皬缇や綋鈥�Pst鈪�/Rsa鈪犲ぇ榛勯奔鍩哄洜缁勪唬琛ㄦ€NA鐗囨, 骞跺姞涓奀y3鑽у厜鏍囪, reference DNA涓篊y5鑽у厜鏍囪銆傛潅浜ゅ悗杩涜鑺墖鎵弿(鍥� 3A)鍜屾暟鎹彁鍙栥€傛瘡寮犺姱鐗囩粡杩囧綊涓€鍖栧鐞嗗悗, 閫氳繃鐢ㄦā绯奀-鍧囧€艰仛绫诲垎鏋愭硶鑾峰緱0/1鐭╅樀銆傛牴鎹绠楁墍寰楃殑p鍊艰幏寰楁暎鐐瑰浘(鍥� 2B)銆傛暎鐐瑰浘涓粦鑹蹭负鏃犲樊寮備綅鐐�, 绾㈣壊涓哄樊寮傛樉钁椾綅鐐�(P鈮�0.05), 鍚勪綅鐐归泦涓湪瀵硅绾块檮杩�, 鍋忕瀵硅绾胯秺澶ц秺瀹规槗鍛堢幇绾㈣壊銆備笂杩板樊寮備綅鐐逛腑鍙湁18涓狣ArT鍊欓€夋爣璁板湪鈥滄瀬绔ぇ缇や綋鈥濅笌鈥滄瀬绔皬缇や綋鈥濅腑鑱氱被缁撴灉绋冲畾, 涓旂粍闂�P < 0.01(琛� 2), 鍏朵腑17涓负鈥滄瀬绔ぇ缇や綋鈥滵ArT鍊欓€夋爣璁�, 1涓€滄瀬绔皬缇や綋鈥滵ArT鍊欓€夋爣璁�(琛� 2)銆�
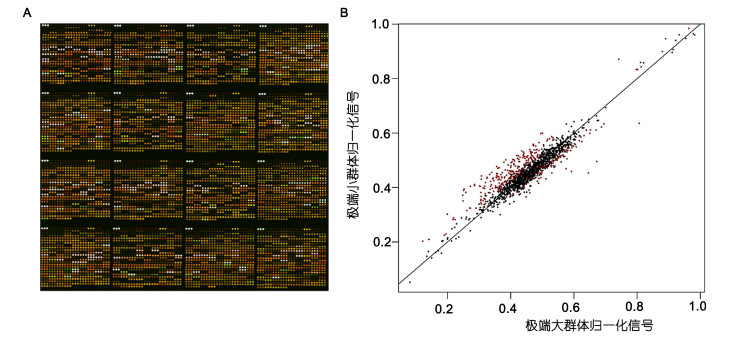 |
| 鍥� 3 x827鏍峰搧鏉備氦缁撴灉(A)鍙婃潅浜ゅ綊涓€鍖栦俊鍙�P鍊兼暎鐐瑰浘(B) Fig. 3 Result of DArT microarray assay of x827 (A) and the P value scatter plot of normalization signals from maximal length group and minimal length group (B) |
| 鍊欓€夋爣璁� | 鏋佺澶х兢浣� | 鏋佺灏忕兢浣� | P鍊� | q鍊� | FDR |
| Rsa鈪�1-05 | 0 | 1 | 9.96E-06 | 0.000493 | 0.000649 |
| Rsa鈪�1-23 | 0 | 1 | 0.000119 | 0.002948 | 0.003886 |
| Rsa鈪�4-73 | 0 | 1 | 8.25E-07 | 6.71E-05 | 8.84E-05 |
| Rsa鈪�5-05 | 0 | 1 | 1.02E-06 | 7.72E-05 | 0.000102 |
| Rsa鈪�5-23 | 0 | 1 | 3.8E-05 | 0.001442 | 0.001901 |
| Rsa鈪�8-80 | 0 | 1 | 2.21E-07 | 2.09E-05 | 2.76E-05 |
| Rsa鈪�9-38 | 1 | 0 | 0.000111 | 0.002806 | 0.003699 |
| Rsa鈪�12-24 | 0 | 1 | 0.000281 | 0.005217 | 0.006877 |
| Rsa鈪�12-18 | 0 | 1 | 2.57E-07 | 2.25E-05 | 2.96E-05 |
| Rsa鈪�13-24 | 0 | 1 | 4.16E-06 | 0.000263 | 0.000347 |
| Rsa鈪�14-11 | 0 | 1 | 9.81E-06 | 0.000493 | 0.000649 |
| Rsa鈪�14-18 | 0 | 1 | 6.09E-08 | 7.69E-06 | 1.01E-05 |
| Rsa鈪�14-73 | 0 | 1 | 0.001214 | 0.013949 | 0.018387 |
| Rsa鈪�15-17 | 0 | 1 | 9.26E-05 | 0.002478 | 0.003267 |
| Rsa鈪�16-85 | 0 | 1 | 7.01E-06 | 0.000399 | 0.000527 |
| Rsa鈪�16-17 | 0 | 1 | 3.09E-06 | 0.000218 | 0.000288 |
| Rsa鈪�14-79 | 0 | 1 | 7.02E-06 | 0.000399 | 0.000527 |
| Rsa鈪�18-18 | 0 | 1 | 8.56E-06 | 0.000464 | 0.000612 |
涓洪獙璇佹墍绛涢€塂ArT鍊欓€夋爣璁扮殑鏈夋晥鎬�, 鍙堥噸鏂版寫閫変簡娴欐睙璞″北娓咕缃戠鍏绘畺鐨勨€滀笢娴�1鍙封€滷6浠eぇ榛勯奔(1榫�) 177灏惧ぇ榛勯奔, 杩欎釜缇や綋浣撻暱缁忚繃Shapiro-Willie杩囩▼杩涜姝f€佸垎甯冩楠�, 涔熺鍚堟鎬佸垎甯�(P > 0.05)銆備粠妫€楠岀粨鏋滃彲浠ョ湅鍒�, Rsa鈪�1-23绛�8涓狣ArT鏍囪浠嶄笌浣撻暱鎬х姸绱у瘑鐩稿叧(P < 0.01) (琛� 3)銆備负浜嗚繘涓€姝ョ‘瀹氱瓫閫夎幏寰楃殑澶ч粍楸糄ArT鍊欓€夋爣璁�, 灏嗕笂杩板€欓€夋爣璁拌繘琛屾祴搴�, 娴嬪簭缁撴灉鐢˙LAST2GO杞欢杩涜Blastn鍒嗘瀽銆傜粨鏋滄樉绀烘祴瀹氱殑搴忓垪涓�6涓负宸茬煡搴忓垪, 2涓负鏈煡搴忓垪(琛� 3)銆�
| 鍊欓€夋爣璁� | 鐧诲綍鍙� | 搴忓垪闀垮害 | 搴忓垪鎻忚堪 | 鏋佺澶х兢浣� | 鏋佺灏忕兢浣� | P鍊� | q鍊� | FDR |
| Rsa鈪�1-23 | KX353586 | 445 | Homo sapiens phosphatase complete cds | 0 | 1 | 0.0001422 | 0.0005448 | 0.0024499 |
| Rsa鈪�4-73 | KX353587 | 789 | Homo sapiens genomic chromosome 11q clone: rp11-complete sequences |
0 | 1 | 1.22E-06 | 1.92E-05 | 8.64E-05 |
| Rsa鈪�8-80 | KX353588 | 224 | Epinephelus coioides complement component c3 complete cds |
0 | 1 | 1.46E-06 | 2.12E-05 | 9.55E-05 |
| Rsa鈪�9-38 | KX353589 | 1042 | 鈥擭A鈥� | 1 | 0 | 0.000195 | 0.000666 | 0.00299 |
| Rsa鈪�14-11 | KX353590 | 669 | Homo sapiens serine hydrolase-like 2 transcript variant non-coding RNA |
0 | 1 | 4.03E-06 | 0.000200 | 0.000195 |
| Rsa鈪�16-85 | KX353591 | 342 | 鈥擭A鈥� | 0 | 1 | 2.62E-06 | 0.000113 | 0.000508 |
| Rsa鈪�14-79 | KX353592 | 450 | glucan-beta-glucosidase | 0 | 1 | 9.35E-06 | 8.47E-05 | 0.000380 |
| Rsa鈪�18-18 | KX353593 | 357 | serine threonine protein kinase | 0 | 1 | 8.61E-06 | 8.01E-05 | 0.000360 |
| 聽聽聽聽娉�: 鈥淣A鈥濊〃绀烘病鏈夋敞閲� | ||||||||
浼犵粺閫夎偛闇€瑕佺粡鍘嗗涓敓鍛藉懆鏈熸墠鑳藉垎绂诲嚭绋冲畾閬椾紶鐨勭粡娴庢€х姸銆傞殢鐫€鍒嗗瓙鐢熺墿瀛︽妧鏈殑杩呯寷鍙戝睍, 杩愮敤DNA鍒嗗瓙鏍囪鎶€鏈繘琛岄€夎偛, 浠庡垎瀛愭按骞崇爺绌朵笌姘翠骇鍔ㄧ墿浼樿壇缁忔祹鎬х姸鐩歌繛閿佺殑鍒嗗瓙鏍囪鏋佸ぇ鍦扮缉鐭簡閫夎偛鏃堕棿(鍒樿搐寰风瓑, 2013)銆傜洰鍓�, SSR鎶€鏈父鐢ㄤ簬瀵绘壘涓庣敓闀裤€佹姉閫嗙瓑鎬х姸绱у瘑杩為攣鎴栫浉鍏虫爣璁般€傚妯婁匠浣崇瓑(2009)鍏宠仈鍒嗘瀽寰楀埌7涓井鍗槦浣嶇偣涓庝綋閲嶃€佷綋闀垮拰浣撻珮鏄捐憲鐩稿叧(P < 0.05)鎴栨瀬鏄捐憲鐩稿叧(P < 0.01)銆�Yi绛�(2015)浠�100涓猄SR鏍囪涓壘鍒�8涓熀鍥犲骇涓�9涓熀鍥犲瀷涓庨硿楸肩敓闀挎€х姸(浣撻噸銆佷綋闀垮拰浣撻珮)鐩稿叧銆傜爺绌惰€呬篃璇曞浘閫氳繃鐢熼暱銆佹姉閫嗙瓑鎬х姸绱у瘑杩為攣鎴栫浉鍏虫爣璁版寚瀵煎ぇ榛勯奔閬椾紶鑲茬瀹炶返銆�楂樺浗寮虹瓑(2010)杩涜浜嗗ぇ榛勯奔鑰愪綆娓╂爣璁扮殑绛涢€�, 鎵惧埌涓€涓爣璁�(LYC0002)鍙兘涓庤€愪綆娓╂湁鍏炽€傚垬璐ゅ痉绛夐噰鐢ㄥ井鍗槦鏍囪鎶€鏈壌瀹氫簡LYC0088鍜孡YC0143涓庡ぇ榛勯奔涓嶅悓瀹剁郴鍜岀兢浣撶敓闀挎€х姸绱у瘑鐩稿叧(鍒樿搐寰风瓑, 2012, 2013; 鍙跺崕绛�, 2014)銆�钖涜壇涔夌瓑(2008)鐮旂┒琛ㄦ槑澶ч粍楸艰倢鑲夌敓闀挎姂鍒剁礌鍩哄洜3鈥欑闈炵紪鐮佸尯寰崼鏄熷簭鍒楀鎬佹€т笌澶ч粍楸间綋闀裤€佷綋璐ㄩ噺鏃犵浉鍏虫€с€�Ni绛�(2012)鍦ㄦ禉姹熷吇娈栧ぇ榛勯奔鐢熼暱鍩哄洜鍐呭惈瀛�1鐨�196浣峉NP(Single nucleotide polymorphysim)涓庝綋闀垮拰浣撻珮鐩稿叧, 鍦ㄤ袱涓兢浣撳ぇ榛勯奔鐢熼暱鍩哄洜鍐呭惈瀛�2鐨�692浣峉NP涓庝綋閲嶅叏闀挎樉钁楃浉鍏炽€傛湰鐮旂┒涓娆¢噰鐢―ArT鎶€鏈壌瀹氫簡8涓ぇ榛勯奔浣撻暱鐩稿叧DArT鏍囪, 鍏朵腑7涓负鈥滄瀬绔ぇ缇や綋鈥滵ArT鍊欓€夋爣璁�, 1涓€滄瀬绔皬缇や綋鈥滵ArT鍊欓€夋爣璁般€�
涓€鑸湁涓ょ鏂规硶绛涢€夊拰閴村畾姘翠骇鍔ㄧ墿鐩爣鎬х姸鐩稿叧鑱旂殑鍒嗗瓙鏍囪, 涓€绉嶆槸鍒嗙缇や綋鏍囪鍏宠仈鍒嗘瀽娉�, 璇ユ柟娉曞彲蹇€熻幏寰椾笌鐩殑鎬х姸杩為攣鐨勫垎瀛愭爣璁�, 缂虹偣鏄伒鏁忓害鍜岀簿纭害閮借緝浣�; 鍙︿竴绉嶄负闅忔満閫夋嫨缇や綋鏍囪鍏宠仈鍒嗘瀽娉�, 璇ユ柟娉曞彲鍏ㄩ潰鍒嗘瀽鎵€閫夌敤鐨勬爣璁�, 鍑嗙‘搴﹀拰绮剧‘搴﹁緝濂�, 缂虹偣鏄渶瑕佹娴嬬殑鏍锋湰閲忓ぇ鍜屽垎鏋愯垂鐢ㄨ緝楂�(妯婁匠浣崇瓑, 2009)銆傛湰鐮旂┒閲囩敤鍒嗙缇や綋鏍囪鍏宠仈鏋愭硶杩涜鍒濇绛涢€�, 鏍规嵁鈥滀笢娴�1鍙封€濆ぇ榛勯奔浣撻暱鏁版嵁鎸戝嚭鈥滄瀬绔ぇ缇や綋鈥濆拰鈥滄瀬绔皬缇や綋鈥�, 浠庝袱涓兢浣撲腑鑾峰緱18涓笌浣撻暱鐩稿叧DArT鍊欓€夋爣璁般€傞殢鍚庢柊缇や綋涓獙璇佺‘璁�Rsa鈪�1-23绛�8涓狣ArT鏍囪浠嶄笌浣撻暱鎬х姸绱у瘑鐩稿叧銆傜敱浜庝粛涓嶈兘淇濊瘉鏈爺绌朵腑鎵€绛涢€夊埌鐨勬爣璁板湪鍏跺畠缇や綋閫傜敤, 鍥犺€屽湪鍚庣画鐮旂┒涓皻闇€鎵╁ぇ缇や綋鐨勮妯″拰绫诲瀷, 杩涜澶氭柟姣旇緝杩涜杩涗竴姝ラ獙璇�, 涓轰笅涓€姝ュ熀鍥犺緟鍔╄偛绉嶃€佽繘涓€姝ユ彁楂樻€х姸閫夋嫨鐨勫噯纭€ф彁渚涗緷鎹€�
4 缁撹鏈爺绌朵粠Pst鈪�/Rsa鈪犲熀鍥犵粍浠h〃DNA鐗囨鏂囧簱涓墿澧炶幏寰�3360涓熀鍥犵粍浠h〃鎬NA鐗囨, 鍒跺澶ч粍楸间綋闀跨浉鍏冲鏍锋€ц姱鐗囥€傛潅浜や俊鍙疯浆鎹负0/1鐭╅樀骞惰繘琛岀粺璁″垎鏋愮瓫閫�, 鍒濇鑾峰緱18涓ぇ榛勯奔浣撻暱鐩稿叧DArT鍊欓€夋爣璁般€備箣鍚庣粡杩囬獙璇佽〃鏄庝粛鏈�8涓狣ArT鏍囪涓庢柊缇や綋鐨勪綋闀跨浉鍏炽€傛湰鐮旂┒鎴愬姛搴旂敤DArT鎶€鏈瓫閫夊埌涓庝綋闀跨浉鍏虫爣璁�, 璇ユ柟娉曚篃鍙互搴旂敤浜庡叾瀹冪敓闀跨浉鍏虫€х姸鏍囪绛涢€夈€傚悓鏃舵湰鐮旂┒鎴愭灉鍙洿鎺ユ寚瀵煎ぇ榛勯奔浜哄伐閫夎偛宸ヤ綔; 涔熷彲浣滀负杩涗竴姝ョ爺绌跺ぇ榛勯奔鐢熼暱鐩稿叧鐨勯仐浼犺繛閿佸浘璋辨彁渚涘熀纭€, 骞跺鍏跺畠姘翠骇缁忔祹鍔ㄧ墿鐨勯仐浼犺偛绉嶇殑寤虹珛鎻愪緵鐞嗚渚濇嵁涓庡疄璺靛弬鑰冦€�
| 鐜嬪織鍕�, 鐜嬭壓纾�, 鏋楀埄姘�, 绛�, 2002. 绂忓缓瀹樹簳娲嬪ぇ榛勯奔AFLP鎸囩汗澶氭€佹€х殑鐮旂┒. 涓浗姘翠骇绉戝, 9 (3) : 198鈥�202 |
| 鐜嬫檽娓�, 鐜嬪織鍕�, 璋腑鍥�, 绛�, 2008. 澶ч粍楸�(鈾€)涓庨(鈾�)鏉備氦鐨勯仐浼犲垎鏋�. 姘翠骇瀛︽姤, 32 (1) : 51鈥�57 |
| 鍙跺崕, 鍒樻磱, 鍒樿搐寰�, 绛�, 2014. 澶ч粍楸煎井鍗槦鏍囪涓庣敓闀挎€х姸鐨勭浉鍏冲垎鏋�. 瑗垮崡澶у瀛︽姤:鑷劧绉戝鐗�, 36 (3) : 27鈥�33 |
| 瀹佸渤, 2007.澶ч粍楸奸仐浼犺繛閿佸浘璋辩殑鏋勫缓.闆嗙編:闆嗙編澶у纭曞+瀛︿綅璁烘枃, 23-52 http://cdmd.cnki.com.cn/article/cdmd-10390-2007155211.htm |
| 鍒樿搐寰�, 闊︿俊閿�, 钄℃槑澶�, 绛�, 2012. 澶ч粍楸�22涓井鍗槦鏍囪鍦‵1瀹剁郴涓殑鍒嗙鏂瑰紡鍙婁笌鐢熼暱鎬х姸鐨勭浉鍏冲垎鏋�. 姘翠骇瀛︽姤, 36 (9) : 1322鈥�1330 |
| 鍒樿搐寰�, 闅嬬彮鑹�, 鐜嬪織鍕�, 绛�, 2013. 澶ч粍楸煎揩闀跨浉鍏冲井鍗槦鏍囪鐨勭瓫閫変笌楠岃瘉. 姘寸敓鐢熺墿瀛︽姤, 37 (6) : 1036鈥�1043 |
| 鏉庢槑浜�, 寮犳捣鐞�, 钖涜壇涔�, 绛�, 2003. 缃戠鍏绘畺澶ч粍楸奸仐浼犲鏍锋€х殑鍚屽伐閰跺拰RAPD鍒嗘瀽. 涓浗姘翠骇绉戝, 10 (6) : 523鈥�525 |
| 鏉庨箯椋�, 鍛ㄦ案涓�, 寰愭眽绁�, 2008. 澶ч粍楸笺€侀楸煎強缇庡浗绾㈤奔绾跨矑浣揇NA鐨凜yt b鍩哄洜搴忓垪姣旇緝. 鍗楁柟姘翠骇, 4 (3) : 43鈥�47 |
| 鑽f檾濠�, 闄堝己, 鍙查洦绾�, 绛�, 2014. 鍩轰簬DArT鏍囪鐨勪笁鐤f瀛愯煿鍦扮悊绉嶇兢閬椾紶澶氭牱鎬у垎鏋�. 鐢熺墿瀛︽潅蹇�, 31 (2) : 18鈥�21 |
| 濮氬悍, 鐜嬫枃绛�, 鑰挎姊�, 绛�, 2008. 澶ч粍楸肩揣瀵嗚繛閿佄�-鍜屛�-鐝犺泲鐧藉熀鍥犻棿搴忓垪鍔熻兘鍒嗘瀽. 姘寸敓鐢熺墿瀛︽姤, 32 (3) : 413鈥�416 |
| 楂樺浗寮�, 甯哥帀姊�, 闊╁惎闇�, 绛�, 2010. 澶ч粍楸艰€愪綆娓╂€х姸鐩稿叧寰崼鏄熸爣璁扮殑绛涢€�. 閬椾紶, 32 (3) : 248鈥�253 |
| 妯婁匠浣�, 鐧戒繆鏉�, 鏉庡皬鎱�, 绛�, 2009. 澶у彛榛戦矆鐢熼暱鎬х姸鐨勫井鍗槦DNA鏍囪绛涢€�. 閬椾紶, 31 (5) : 515鈥�522 |
| 钖涜壇涔�, 瀛欏崌, 鑲栫珷濂�, 绛�, 2008. 澶ч粍楸艰倢鑲夌敓闀挎姂鍒剁礌鍩哄洜寰崼鏄熷簭鍒楀鎬佹€у垎鏋�. 涓浗鐢熺墿鍖栧涓庡垎瀛愮敓鐗╁鎶�, 24 (10) : 980鈥�985 |
| 榄忎笢鏃�, 妤煎厑涓�, 瀛欐晥鏂�, 绛�, 2001. 椴ら奔寰崼鏄熷垎瀛愭爣璁扮殑绛涢€�. 鍔ㄧ墿瀛︾爺绌�, 22 (3) : 238鈥�241 |
| Chang Y M, Ding L, Wang W W, et al, 2009. Isolation and characterization of 11 microsatellite markers for the large yellow croaker, Pseudosciaena crocea. Conservation Genetics, 10 (5) : 1405鈥�1408 DOI:10.1007/s10592-008-9708-9 |
| Gao G Q, Chang Y M, Han Q X, et al, 2010. Screening of microsatellite markers associated with cold tolerance of large yellow croaker (Pseudosciaena crocea R. ). Hereditas, 32 (3) : 248鈥�253 |
| Guo W, Wang Z Y, Wang Y L, et al, 2005. Isolation and characterization of six microsatellite markers in the large yellow croaker (Pseudosciaena crocea Richardson). Molecular Ecology Notes, 5 (2) : 369鈥�371 DOI:10.1111/men.2005.5.issue-2 |
| Hackl E, Konrad-K枚szler M, Kilian A, et al, 2010. Phage-type specific markers identified by Diversity Arrays Technology(DArT) analysis of Salmonella enterica ssp. enterica serovars Enteritidis and Typhimurium. J Microbiol Meth, 80 (1) : 100鈥�105 |
| Jaccoud D, Peng K M, Feinstein D, et al, 2001. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res, 29 (4) : e25 DOI:10.1093/nar/29.4.e25 |
| Ni J, You F, Xu J H, et al, 2012. Single nucleotide polymorphisms in intron 1 and intron 2 of Larimichthys crocea growth hormone gene are correlated with growth traits. Chin J Oceanol Limnol, 30 (2) : 279鈥�285 DOI:10.1007/s00343-012-1078-y |
| S谩nchez-Sevilla J F, Horvath A, Botella M A, et al, 2015. Diversity Arrays Technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the Octoploid cultivated strawberry (Fragaria脳ananassa). PLoS One, 10 (12) : e0144960 DOI:10.1371/journal.pone.0144960 |
| Schouten H J, van de Weg W E, Carling J, et al, 2012. Diversity arrays technology (DArT) markers in apple for genetic linkage maps. Mol Breed, 29 (3) : 645鈥�660 DOI:10.1007/s11032-011-9579-5 |
| Wenzl P, Carling J, Kudrna D, et al, 2004. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci U S A, 101 (26) : 9915鈥�9920 DOI:10.1073/pnas.0401076101 |
| Ye H, Liu Y, Liu X D, et al, 2014. Genetic mapping and QTL analysis of growth traits in the large yellow croaker Larimichthys crocea. Mar Biotechnol, 16 (6) : 729鈥�738 DOI:10.1007/s10126-014-9590-z |
| Yi T L, Fang L, Liang X F, et al, 2015. Characterization of microsatellite markers and their correlations with growth traits in Mandarin fish (Siniperca chuatsi). Genet Mol Res, 14 (3) : 8926鈥�8934 DOI:10.4238/2015.August.7.1 |
 2016, Vol. 47
2016, Vol. 47


