文章信息
- 陈楠生. 2020.
- CHEN Nan-sheng. 2020.
- 有害藻华的宏条形码分析:机会与挑战
- Metabarcoding analysis of harmful algal blooms: Opportunities and challenges
- 海洋科学, 44(7): 116-134
- Marine Sciences, 44(7): 116-134.
- http://dx.doi.org/10.11759/hykx20200310001
-
文章历史
- 收稿日期:2020-03-10
- 修回日期:2020-04-13
2. 青岛海洋科学与技术试点国家实验室 海洋生态与环境科学功能实验室, 山东 青岛 266237;
3. 中国科学院海洋大科学研究中心, 山东 青岛 266071
2. Marine Ecology and Environmental Science Laboratory, Pilot National Laboratory for Marine Science and Technology(Qingdao), Qingdao 266237, China;
3. Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
有害藻华(harmful algal blooms)是由于藻类快速繁殖和累积造成的自然生态现象, 可能具有灾难性的生态或经济后果, 比如导致海洋动、植物死亡、海洋食品污染、海洋食物链的改变, 以及旅游资源的破坏等[1-2]。近年来, 国内外赤潮发展具有新特点, 包括暴发规模加大、持续时间更长、致灾效应加重和全球扩张明显[3]。我国海岸线长, 近岸海域面积大, 包括渤海、黄海、东海和南海, 有害藻华是我国海洋生态系统中最重要的问题之一[1]。对于引发有害藻华的物种, 应用传统的基于形态分类的研究方法, 积累了大量的数据。然而基于形态分类的研究方法有很大的局限性。首先, 需要研究人员具有专业经验, 来准确分辨不同藻类物种; 其次, 藻类细胞在分析过程中的形态稳定性的不同可能造成结果的偏差; 另外, 细胞太小的微微藻往往会被忽略; 还有, 形态相似的隐存种得不到准确区分[4-6]。
分子分析方法的引入大大加强了我们对包括有害藻华物种在内的浮游植物生物多样性的理解[7]。早在20多年前, 美国和法国科学家分别通过克隆测序叶绿体16S rDNA和核糖体18S rDNA序列鉴定到海水中存在多种真核浮游植物[5, 8-9](图 1)。随后, 18S rDNA被作为分子标记研究自然海域浮游植物的组成, 并发现两类与甲藻进化关系较近的生物类群, 即海洋alveolateⅠ和alveolateⅡ[10-11], 并确认了AlveolateⅠ和AlveolateⅡ都是寄生性甲藻[12]。
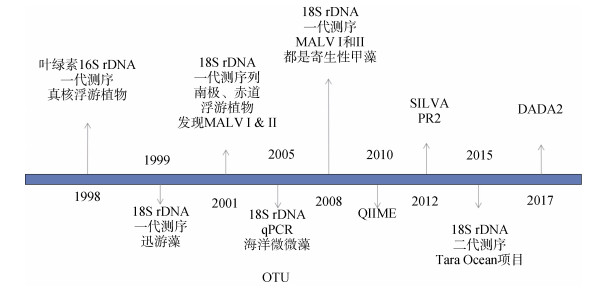 |
| 图 1 宏条形码分析的里程碑事件 Fig. 1 Milestones in metabarcoding analysis |
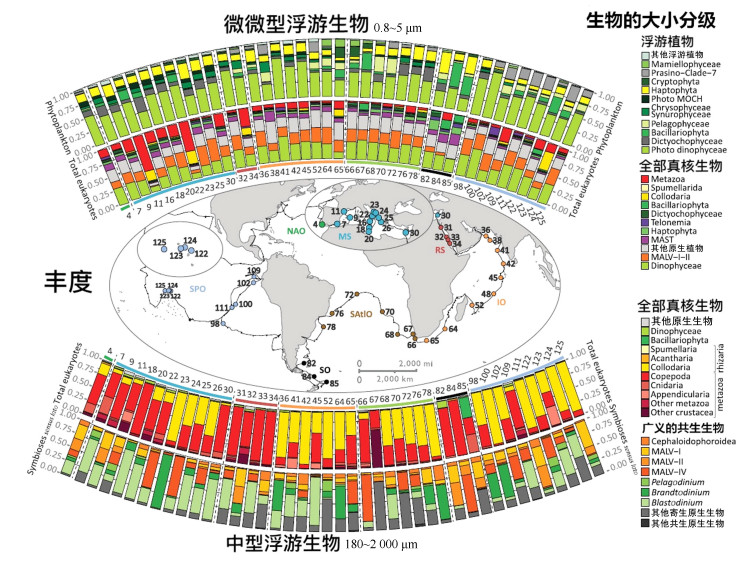
这些基于DNA分子标记扩增、测序研究野外样本的方法迅速显示出强大的潜力, 并逐步演变成为宏条形码分析[13], 即针对包括很多物种的自然样本, 而不是单一物种为研究对象的基因组研究。宏条形码分析已广泛应用于包括有害藻华在内的浮游植物的生态学研究[14]。宏条形码分析策略与基于全基因组测序的宏基因组学分析(metagenomics analysis)相辅相成, 宏条形码分析关注物种组成及相对丰度, 宏基因组学则重点关注基因功能。2015年美国《科学》杂志发表了系列利用宏条形码分析的文章, 报道Tara Oceans国际合作项目的研究成果(图 2)[15]。通过对采集到的334个浮游植物样本针对18S rDNA V9进行扩增和高通量测序, 得到了150 000个可操作分类单元(operational taxonomy units, OTUs), 其中1/3未能比对到已知物种, 说明海洋中存在大量的尚未得到描述的真核物种。Tara Oceans数据的分析结果揭示了藻类物种的全球分布格局, 包括藻类物种的生物地理学[16-22]、季节性变化规律[23-24]和共生性[25]。另外一个有代表性的宏条码研究是由国际海洋采样日联盟(Ocean Sampling Day Consortium)发起的国际合作项目[26-27]。这个由欧盟资助的微生物B3项目发起的国际组织意在通过国际合作, 在同时间获取海洋样本, 完成测序分析。迄今, 海洋采样日联盟已经协同完成了多个重大项目, 比如, 通过对全球141个采样位点样本的同时采样, 和18S rDNA V4序列的测序, 首次完成了大规模海洋绿藻的全球分布分析[26]。近年来, 随着DNA测序技术在海洋生态领域的应用, DNA测序正在代替传统技术, 逐步被应用于监测海洋生态状况[28]。另外, 国际合作项目全球微生物组计划(Earth Microbiome Project)也展开了较多的海洋浮游植物的宏条形码分析[29]。
宏条形码分析方法引入我国以后, 促进了针对各海域的有害藻华研究[30-33]。特别是近年来, 宏条形码研究越来越多, 包括对渤海[34-38]、黄海、东海[39-40]、南海[41-43]、胶州湾[4, 44]、北部湾、长江口、珠江口[45-46]等的研究, 取得了显著进展。本综述系统介绍这些进展, 并分析存在的机会与挑战。
1 宏条形码分析技术 1.1 条形码的选择宏条形码分析通用的条形码包括核糖体rRNA基因18S rDNA、28S rDNA、ITS序列、叶绿体基因rbcL和16S rDNA[47]和线粒体基因CO1等。这些通用分子标记的最大优势在于它们的通用性, 即可以通过设计通用PCR扩增引物进行扩增、测序, 并且可以用于分析绝大多数的浮游植物, 包括有害藻华物种。早期的宏条形码分析是通过克隆、测序策略完成的, 即扩增全长或局部的通用分子标记, 克隆, 然后挑选单克隆, 通过第一代DNA测序(即桑格测序)技术读取每一条条形码的序列。随着第二代DNA测序技术的出现, 高通量、低成本的第二代DNA测序技术逐步取代低通量、高成本的第一代DNA测序技术, 变成宏条形码主流测序技术。由于第二代DNA测序技术的读长比较短, 因此基于第二代DNA测序技术的宏条形码分析往往针对上述通用分子标记的局部, 比如, 18S rDNA的V4区域[4]或V9区域[15], 28S rDNA的D1-D2区域[48]等特异性条形码。目前, 第二代DNA测序技术是开展宏条形码研究的主流, 但是, 由于二代DNA测序得到的DNA片段往往不足以分辨不同物种, 基于第二代DNA测序的宏条形码分析不能充分回答很多问题。随着第三代单分子DNA测序技术的日益成熟, 特别是基于PacBio测序平台的测序通量的不断增加, 成本逐步下降, 越来越多的人开始尝试将第三代DNA测序技术应用于宏条形码分析[49]。
1.2 宏条码分析方法:从OTU分析到ASV分析很长一段时间以来, 宏条形码分析的通常分析思路是通过分子标记(即条形码)扩增和测序后, 对测序结果进行聚类分析, 生成可操作分类单元(OTU)。每一个可操作分类单元OTU包括样本中相差很小(通常在3%以内)的序列。OTU分析方法被广泛用于分析宏条形码研究项目。通常, 人们期待OTU具有如下两方面的功能:其一, 把针对某种分子标记的环境样本的高通量测序结果转换成物种分类信息, 并且不同的相似性对应不同的分类单元; 其二, 通过把相似的序列聚类起来, 从而减少测序错误带来的副作用, 降低对生物多样性分析和物种组成分析的负面影响[50]。然而, OTU并不能够很好地执行上面的两个功能。OTU并没有具体的分类信息, 不能与物种进行一一对应。而且, 模拟研究发现OTU聚类分析得到的OTU数目往往远远多于物种数目, 表明OTU聚类分析具有很高的假阳性[50]。同时, 由于OTU聚类分析忽视了现代测序结果中的质量信息, 浪费了区分更精细的序列多样性的可能性, 人为限制了OTU聚类分析的分辨率, 表明OTU聚类分析还具有很高的假阴性[51-52]。
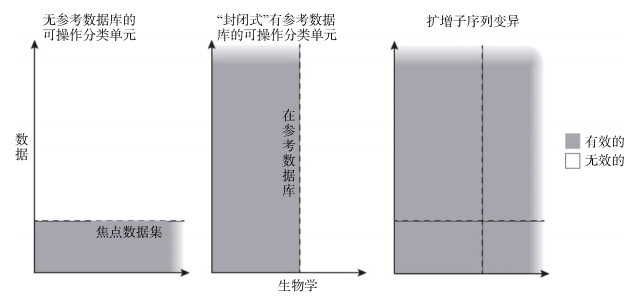
OTU聚类分析的另外一个严重的弊端是分析结果不能用于比较不同样本的物种组成。无参考数据库的OTUs(de novo OTUs)分析结果非常依赖样本的组成(图 3), 因此, 两个不同样本的OTU聚类分析结果不能进行比较。为了支持不同样本聚类分析结果的可比性, 人们提出了“封闭式”有参OTUs(closed- reference OTUs)的概念, 即在聚类过程中, 根据相似性, 序列都围绕参考数据库中的参考序列进行聚类, 得到OTUs。因此, 只要利用相同的参考数据库, 分析得到的OTUs是可以比较的。这个分析思路的局限是, 参考数据库中不存在但是样本中存在的数据就不能得到聚类(图 3)。因此, 封闭式有参OTUs分析受到参考数据库的局限[50]。并且, 参考数据库的任何变化比如数据库升级都会影响分析结果。
为了解决上述OTU相关的问题, 人们提出了基于扩增子序列变异(amplicon sequence variant, ASV)概念。ASV算法的基本假设是真实的DNA序列在测序结果中出现的频率远远高于测序误差, 不依赖阈值区分不同的ASV[50]。因此, 来自不同样本的ASV序列具有可比性[50]。
基于这个概念的宏条形码分析具有既不依赖参考数据库, 也不依赖相似性阈值的优点。因此, 与基于OTU概念的分析方法相比, 基于ASV的宏条形码分析方法具有很高的优越性, 可以捕获样本中所有的变异, 并且可以用于比较分析不同样本的物种组成(图 3)。由于基于ASV的分析更加准确, 分析结果可以用于比较不同样本的物种组成, 可以重复使用, 并且更加全面, 基于OTU的宏条形码分析方法在逐步被基于ASV的方法替代[50, 53]。常用的用于分析ASV的方法包括DADA2[51], Deblur[54]和denoise[55]。
DADA算法, 即不同扩增子去噪算法(Divisive Amplicon Denoising Algorithm), 最早发表于2012年[52]。DADA是在AmpliconNoise算法[55]的基础上开发出来的。DADA的优势在于不需要训练数据集, 充分利用序列丰度信息, 通过误差建模, 评估误差参数, 最后推测出样本测序结果中的序列类型。2016年DADA2发表, 特别增加了针对Illumina扩增子的测序质量模型, 并开发了R软件包, 全部分析可以在R环境下运行, 包括过滤、去重复、推测序列信息、鉴定嵌合体、结合双端测序结果等[51]。最近, 我们成功利用DADA2方法分析胶州湾2019年冬季航次采集的样本, 分析发现了大量浮游植物, 其中包括首次在胶州湾发现的有害藻华物种, 以及大量的寄生性甲藻, 并评估了浮游植物之间的互作情况[4]。
2 我国海域有害藻华物种的宏条形码分析进展 2.1 近岸海域的宏条形码分析渤海是我国唯一的内海, 具有半封闭性, 包括辽东湾、渤海湾和莱州湾, 是一个重要的产卵场和索饵场(图 4)[56]。由于陆生污染物排放和富营养化, 赤潮和褐潮暴发频率和强度均呈上升趋势[56-57]。基于形态学的镜检的研究发现渤海代表性有害藻华物种, 包括夜光藻(Noctiluca scintillans)、原甲藻(Prorocentrum sp.)、血红哈卡藻(Akashiwo sanguinea)[58], 丹麦细柱藻(Leptocylindrus danicus)[59]、柔软拟菱形藻(Pseudo-nitaschia delicatissima)[59]、浮动弯角藻(Eucampia zoodiacus)[59]、中肋骨条藻(Skeletonema costatum)[59]、诺氏海链藻(Thalassiosira nordenskioldi)[59]、旋链角毛藻(Chaetoceros curvisetus)[59]、柔弱几内亚藻(Guinardia delicatula)[60]、多形微眼藻(Minutocellus polymorphus)[61]和有害甲藻(Stoeckeria algicida)[36, 62-63]。基于扩增测序18S rDNA V4的宏条形码研究证实褐潮的致灾藻华物种是抑食金球藻(Aureococcus anophagefferens)[64-65]。通过对沉积物样本进行18S rDNA V9区扩增测序, 发现了抑食金球藻是一个本地种, 并且是一个在全世界范围内广泛分布的有害藻华物种[66]。除此之外, 宏条形码研究还发现渤海还存在很多丰度较低但是又具有暴发潜力的有害藻华物种[34, 36, 38, 67-69]。
黄海是一个陆缘海(marginal sea), 具有比较复杂的海流包括黄海冷水团(Huanghai Cold Water Mass, HCWM), 黄海暖流(Huanghai Warm Current, HWC), 以及长江冲淡水(Changjiang River Diluted Water, CDW)(图 4)[70]。苏北浅滩的浮游植物以硅藻为主[71]。孢囊分析发现很多有害藻华物种的孢囊, 包括具刺膝沟藻(Gonyaulax spinifera)和多边舌甲藻(Lingulodinium polyedrum), 链状/塔马亚历山大藻(Alexandrium catenella/tamarense), 微小/相似亚历山大藻(Alexandrium minutum/affine)和链状裸甲藻(Gymnodinium catenatum)[72]。基于18S rDNA V4区的宏条形码比较分析发现南黄海与北黄海海域的绿藻与硅藻具有很高的丰度, 可以支撑海岸带的食物链, 特别是海洋经济贝类的养殖[73]。
位于我国、韩国、日本之间的东海是一个很大的大陆架(continental shelf), 也是一个高产渔场, 其浮游生物受长江冲淡水以及黑潮(Kuroshio Current)的影响比较大[74-75]。东海的长江口附近海域是赤潮暴发最显著的位置[1], 对海洋生态环境和公共卫生产生极大影响[76]。迄今对于东海浮游植物及有害藻华物种的研究以浮游植物色素研究[74]、流式细胞研究[75]和形态观察[77-79]为主。东海有害藻华物种中,东海原甲藻(Prorocentrum donghaiense)[80-83]和米氏凯伦藻(Karenia mikimotoi)[84-85]占主导地位。最近在东海发现了指状卡罗藻(Karlodinium digitatum)[84], 血红哈卡藻的多个隐形种(cryptic species)[85], 以及尖刺拟菱形藻(Pseudo-nitzschia pungens)[86]。厦门岛样本进行18S rDNA V9测序, 发现底栖微型真核生物的多样性比浮游的更高, 不同生境的物种分布模式以及影响分布模式的关键过程不同[39, 87-88]。定鞭藻(haptophyte)是重要的浮游植物, 贡献显著的初级生产力, 也具有很高的生物多样性[89-91]。通过利用定鞭藻特异性引物对18S rDNA V4区的扩增测序及宏基因组分析证实了定鞭藻的分子多样性水平, 并检测到所有定鞭藻的主要分枝, 包括有害藻华物种球形棕囊藻(Phaeocystis globosa)及三个同属的棕囊藻物种(P. cordata, P. johnii, 以及一个由序列ST3500.059代表的物种)[40]。
南海位于西太平洋北部, 是由多个国家围绕的半封闭海域, 也是东南亚最大的半封闭的陆源海[92]。南海的主要有害藻华物种包括夜光藻、球形棕囊藻、中肋骨条藻、锥状斯克里普藻(Scrippsiella trochoidea)、赤潮异弯藻(Heterosigma akashiwo)、海洋卡顿藻(Chattonella marina)和米氏凯伦藻, 有害藻华极大地影响了南海海洋渔业。镜检和形态学分析显示上升流对物种分布具有显著的影响[93]。调查研究系统描述了南海浮游植物的组成和季节性变化, 其中也包括很多有害藻华物种[94-104]。卫星遥感方法也跟踪研究了南海赤潮[105-108], 特别是球形棕囊藻赤潮[109]的形成过程。宏条形码分析揭示了香港水域微微藻生物的多样性, 远高于预期[110]。全长18S rDNA扩增, 克隆测序分析揭示了南海浮游植物的组成和分布格局, 发现甲藻是优势物种[111]。基于ITS2的宏条形码分析发现了珊瑚礁共生藻类的多样性[41-43]。
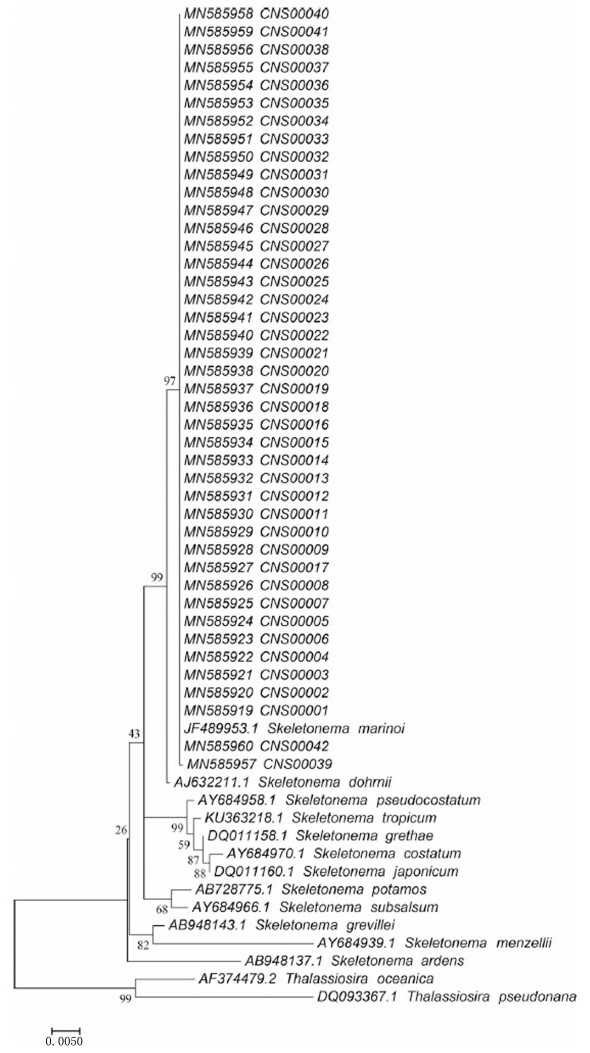
2.2 海湾宏条形码研究胶州湾是黄海西部的一个半封闭性内湾, 海湾与外海可以进行通畅的海水交换, 半交换周期为5天[112]。近30年来, 经济活动加快和人口增长造成了胶州湾显著的富营养化[113-114]。长时序遥感影像分析跟踪胶州湾秋季叶绿素a浓度变化也显示出胶州湾浮游植物群落的巨大变化[115]。胶州湾发现的甲藻孢囊中很多是有害藻华物种[116]。近年来, 胶州湾暖水性浮游植物、甲藻等浮游植物类群的数量呈现升高趋势[117]。胶州湾有记录的有害藻华物种高达68种, 其中有多种在胶州湾引发过赤潮暴发[118]。常见有害藻华物种包括夜光藻[119-120]、浮动弯角藻[121-124]和骨条藻[4]。在挪威海域引发大规模有毒藻华的有害藻华物种里氏金色藻(Chrysochromulina leadbeateri)也在胶州湾发现[125]。历史调查报告中, 胶州湾的骨条藻大多被鉴定为中肋骨条藻[124]。最近我们针对18S rDNA V4的宏条码分析, 加上对分离到的单细胞骨条藻株系的全长18S rDNA测序分析表明, 胶州湾的骨条藻为玛氏骨条藻(Skeletonema marinoi)(图 4)[4]。针对rbcL的宏条形码分析初步揭示了胶州湾浮游植物的生物多样性, 春季代表性物种包括隐藻、定鞭藻和红藻[44]。我们针对18S rDNA的V4片段完成了宏条形码分析, 分析了胶州湾冬季(2019年1月)的浮游植物组成及空间分布格局, 鉴定出胶州湾有28个有害藻华物种, 并发现了胶州湾物种之间的可能互作网络关系(图 5)[4]。
北部湾位于南海西北部的一个半封闭式的海湾, 热带季风气候, 曾经被认为是我国最后“一片干净的海域”和高产渔业基地[126]。进入19世纪90年代, 由于自然与人类活动的双重影响加剧[127], 北部湾海域暴发了一系列赤潮, 包括红海束毛藻(Trichodesmium erythraeum)赤潮和球形棕囊藻赤潮[128-130]。在过去30年里, 北部湾藻华暴发频率、暴发持续时间和暴发规模都有显著上升; 这些赤潮暴发事件不仅影响渔业, 还对附近核电设施造成了巨大的威胁[126]。基于形态学分析在涠洲岛附近鉴定出103种浮游植物, 包括23种有害藻华物种, 其中球形棕囊藻、红海束毛藻、菱形海线藻(Thalassionema nitzschioides)、旋链角毛藻和洛氏角毛藻(Chaetoceros lorenzianus)占优势[129]。利用流式细胞仪对北部湾9个航次的分析揭示了浮游植物在北部湾分布的时空变化[131], 浮游植物在夏季和秋季的丰度较高, 并集中于钦州湾附近以及雷州半岛的西部。镜检发现钦州湾附近的浮游植物也比较丰富, 优势种包括球形棕囊藻和多种硅藻, 季节变化显著[132]。球形棕囊藻囊体在北部湾也呈现出特定的时空分布, 高峰出现在12月到次年3月, 6月囊体很少[133]。迄今利用宏基因组学方法研究北部湾海域有害藻华物种的组成, 以及在有害藻华暴发过程中的动态变化刚刚起步[134]。通过对2014—2015年度样本的18S rDNA V4序列的测序, 重点分析了5种定鞭藻在北部湾北方的钦州湾分布情况, 发现与NO3–N正相关, 并且发现水温是决定球形棕囊藻丰度的主要原因。
2.3 河口宏条形码研究河口往往是多样性程度很高的物理-生物互作生态系统, 由于淡水注入(freshwater discharge)和咸水倒灌(salt water intrusion), 在河口附近盐度和营养盐显示很大的变化, 形成了河流-海域连续带(river ocean continuum)。由于河口附近海域复杂的物理及生物地球化学互作, 使得针对这些水域浮游植物的生长、分布规律, 以及有害藻华暴发的研究都具有很大的挑战。
长江口由于长江径流和泥沙入海, 以及复杂流系结构和特殊地形, 形成了独特的生态系统, 也为赤潮暴发提供了条件[135-137]。长江口及其邻近海域受到长江冲淡水和黑潮分支的双重影响, 有害藻华的形成和演变不仅是陆源污染的“指示器”, 也是反映黑潮分支年际变异的“效应器”[138]。从20世纪60年代到90年代, 人为富营养化(cultural eutrophication)越来越严重[139]。长江口水域有多达68种有害藻华物种, 其中5种有在长江口暴发赤潮的记录[140], 以甲藻为主, 包括东海原甲藻[83]、米氏凯伦藻、亚历山大藻(Alexandrium spp.)和夜光藻[138, 141]。仅次于甲藻的是硅藻, 包括中肋骨条藻[141-143]。中肋骨条藻与夜光藻是常见和多发性赤潮物种, 二者经常形成“混合型”赤潮[144]。长江口内到长江口外的盐度梯度和磷浓度严重影响有害藻华物种的分布[145]。迄今对长江口水域的浮游植物及有害藻华物种的研究以基于形态分析的镜检为主[137, 144, 146-160]。比如, 对长江口24个站位的72个样本进行的形态学鉴定和统计学分析, 鉴定出94个物种, 包括骨条藻、东海原甲藻、米氏凯伦藻。发现不同有害藻华物种引起的不同类型的赤潮在长江口的不同地点同时暴发, 包括一个骨条藻赤潮, 两个东海原甲藻赤潮, 以及一个米氏凯伦藻赤潮。另外发现从河口到离岸海域, 赤潮物种逐步从硅藻向甲藻、裸甲藻转变的趋势[161]。此外近来硅藻有逐步被甲藻替代的趋势[138]。针对18S rDNA的一个1 001 bp片段的宏条形码分析发现长江口水域存在较高的阿米巴藻Amoebophryidae[162]。针对18S rDNA V2-V3的宏条形码研究发现长江口附近海域包括很多的甲藻(特别是裸甲藻Gymnodinium和异冒藻Heterocapsa)和硅藻物种(特别是骨条藻)[163]。迄今为止, 还没有利用宏条形码方法分析长江口中河流-海水连接带的浮游植物的相关研究。
珠江口是一个位于我国南方亚热带河口, 连接南海北部大陆架, 其通量仅次于长江口, 全球排第13位。近30年来, 由于工业发展, 农业活动, 以及城市化, 珠江口富营养化严重[164]。基于藻细胞形态和荧光色素的分析, 发现珠江口的浮游植物中存在多种有害藻华物种, 包括无纹螺沟藻(Gyrodinium instriatum)、中肋骨条藻、浮动弯角藻、海洋原甲藻(Prorocentrum micans)、三角棘原甲藻(Prorocentrum triestinum)、叉角藻(Ceratium furca)、血红哈卡藻、链状裸甲藻、哈曼褐色多沟藻(Pheopolykrikos hartmannii)、双胞旋沟藻(Cochlodinium geminatum)等[165-173]。珠江口浮游植物组成和动态分布的季节性变化, 受包括风暴在内的环境因子的影响[172, 174-175]。迄今珠江口的宏条形码研究不多。基于rbcL的宏条形码的分析展示了珠江口到南海的浮游植物组成, 以及受营养盐和盐度梯度影响的分布方式[46]。珠江口河流-海域连接带的浮游生物在丰水(夏季)和枯水(冬季)之间差异的宏条形码研究创造性地利用了DNA与RNA的不同特性及其反映的不同状态, 比较研究了浮游生物的丰度及活性[176]。
3 机遇与挑战 3.1 宏条形码分析的稳步发展由于DNA测序技术的快速提升, 生物信息学分析方法的迅速跟进, 宏条形码分析也得到了显著的完善, 经历了三个主要发展过程: (1)基于对野外样本的特定分子标记(即条形码)的PCR扩增、分子克隆和第一代DNA测序技术的低通量分析。这个阶段的主要分析方法是将测序结果与已知物种的序列合并在一起, 构建遗传进化树, 分析野外样本的物种组成[10-11, 64, 162]。(2)基于对野外样本的特定分子标记的PCR扩增, 对测序产物进行高通量(第二代DNA测序技术)测序, 利用生物信息学方法进行OTU分析, 解析野外样本的物种组成及相对丰度[36, 44, 163, 176-177]。(3)基于PCR扩增、第二代或第三代DNA测序技术, 利用ASV分析物种组成及相对丰度[4]。
3.2 宏条形码分析的竞争优势与基于形态观察和分类的研究方法相比, 宏条形码研究方法, 具有多方面的显著优势。(1)分辨率比较高, 能够区分隐存种(cryptic species)。在宏条形码分析中, 分子标记中任何差别, 比如只要有单碱基的差别, 都可以通过DNA测序和生物信息学比较检测到。因此分辨率比较高。相比而言, 基于形态观察的分析方法只能够分辨出具有显著区别的形状。因此, 基于形态分析不能分辨的很多不同隐存种, 都依据分子标记信息区分成不同的物种。比如塔玛亚历山大藻“复合种”(“Alexandrium tamarense species complex”)根据分子标记信息被区分为5种藻: A. fundyense、A. mediterraneum、A. tamarense、A. australiense和A. pacificum[178]。(2)适于开展定量分析。在高分辨率基础上, 还可以通过测序结果分析、跟踪每个不同分子标记的数量, 从而能够定量分析包括有害藻华物种(以及所有浮游植物、浮游动物)相对丰度在内的各种多样性指标, 更加准确地计算浮游植物群落的α多样性指数、β多样性指数[179]。(3)宏条形码分析方法比较容易标准化, 因此比较容易学习。相反, 基于形态学的分析方法的掌握需要长时间的专业训练和积累才能够获得对不同物种的认识, 导致针对相同样本, 不同研究者可能得到不同的分析结果, 客观性比较差。(4)比较容易规模化, 采样标准化的宏条形码方法分析我国各个不同海域, 以及相同海域的不同时间点采集的样本, 并且分析结果的可比性比较强。相比而言, 基于形态学的分析方法由于很大程度上依赖研究者的经验和判断能力, 不同项目的研究结果往往可比性较低。因此, 宏条形码分析方法开始应用于研究我国近岸海域的浮游植物组成以及动态变化, 为研究有害藻华的暴发机理提供了很好的支撑。
3.3 宏条形码分析带来的机遇可以期待, 宏条形码分析方法将在我国有害藻华研究中起越来越重要的作用。(1)增加不同项目之间的可比性。迄今为止的基于形态观察的浮游植物和有害藻华物种调查研究虽然取得了巨大的成就, 描绘出我国近岸海域浮游植物和有害藻华物种的组成和分布。然而, 这些组成和分布研究结果大多是定性的。未能实现定量分析的原因有很多, 比如, 采集体积不定量(网采), 研究者的经验和特长不同, 对相同海域的物种鉴定结果可能不一致, 导致不同研究者获得的结果可比性差。基于宏条形码的分析将大大提高不同项目之间的定量可比性。(2)发掘新物种, 研究物种的生物地理学。通过采用规范性采样程序, 开展高通量测序, 宏条形码分析可以发现曾经被忽视的浮游植物物种。很多微微型有害藻华物种, 光学显微镜不能充分发现和区分, 常常被忽略。比如, 北戴河海域的抑食金球藻正是通过宏条形码分析得到证实[64]。抑食金球藻的广泛分布格局也是由宏条码分析方法实现[66]。宏条形码分析发现海水中大量存在的寄生性甲藻[180]。(3)浮游植物-浮游植物互作。海洋生态系统中的浮游植物与其他种类浮游植物、浮游动物、浮游微生物之间都存在着多种多样的相互作用, 包括捕食(predator-prey)、共生(host-symbiont)和寄生(host-parasite)[4, 181]。
3.4 宏条形码分析的挑战宏条形码分析具有极大的竞争力, 将在我国浮游植物研究, 特别是有害藻华物种的分布和丰度的动态研究中起越来越重要的作用。为了确保宏条形码分析方法释放更大的威力, 需要突破两方面的挑战[182]。
第一个挑战是高分辨率分子标记(即条形码)的选择和PCR扩增引物的设计。(1)选择对研究对象具有代表性或特异性, 具有高分辨率的分子标记。宏条形码分析中通常使用的分子标记包括核糖体RNA基因18S rDNA和28S rDNA, ITS, 线粒体基因CO1, 以及叶绿体基因rbcL。使用这些通用分子标记的优势是它们都有通用PCR扩增引物, 并得到充分验证和应用, 具有广泛的可比性。然而, 这些分子标记也有其局限性, 它们往往过于保守, 分辨率有限, 不能够有效区分很多同属物种, 更不能够区分同种的不同遗传株系。除此之外, 这些通用分子标记在基因组中往往以多拷贝存在, 而且拷贝数在不同物种中具有拷贝数多样性, 因此这些分子标记的测序结果不能简单解析其在样本中的相对丰度。最近证据表明, 有些通用分子标记在同一基因组中的多拷贝之间具有序列多样性, 影响了这些分子标记应用价值[183]。为了提升分子标记对不同物种的分辨率, 可以采用第三代DNA测序技术测序样本的全长分子标记。也可以通过比较分析, 开发其他分子标记。比如线粒体、叶绿体或核基因组中的高变异区作为分子标记。通用分子标记18S rDNA的通用扩增引物不能很好扩增定鞭藻的信息, 改用28S rDNA作为分子标记, 扩增到丰富的定鞭藻的信息[48]。(2)设计PCR扩增引物, 降低其偏好性。如果PCR扩增引物具有偏好性, 测序结果就不能够如实反映样本的组成以及动态变化规律。因此, 为了获得较好的结果, 选择适当的分子标记, 设计适当的PCR扩增引物至关重要。
第二个挑战是分子标记数据库的完整性。针对浮游植物和有害藻华物种的分子标记数据库包括SILVA[184]和PR2[185]。在通常的宏条形码分子中, 超过50%(或更多)的测序结果在数据库中没有对应的参考序列, 因此得不到准确注释。随着数据库不断更新, 这种情况在逐步得到改善。不过, 我国海域中有很多特有的浮游植物和有害藻华物种。为了获得对这些物种的覆盖, 我们需要针对中国特异性物种, 充实相应的分子标记, 完善数据库, 促进对我国海域样本宏条形码分析结果的解读。随着这些挑战性问题的解决, 可以预期宏条形码方法将会更加有效应用于有害藻华物种的研究。
此外, 宏条形码分析与其他组学分析技术比如宏基因组学, 宏转录组学, 宏蛋白组学和宏代谢组学具有互补的特性。宏条形码分析集中解析物种组成和相对丰度, 宏基因组学分析基因组功能, 宏转录组学和宏蛋白组学分别从转录水平和蛋白水平分析基因表达以及对环境的响应, 宏代谢组学分析环境一般的代谢产物。这些组学技术结合起来应用将更能发挥其重要性。
| [1] |
Yu R, Lv S H, Liang Y. Harmful algal blooms in the coastal waters of China[J]. Global Ecology and Oceanography of Harful Algal Blooms, 2018, 232: 309-316. |
| [2] |
周名江, 朱明远, 张经. 中国赤潮的发生趋势和研究进展[J]. 生命科学, 2001, 13(2): 54-59. Zhou Mingjiang, Zhu Mingyuan, Zhang Jing. Status of harmful algal blooms and related research activities in China[J]. Chinese Bulletin of Life Sciences, 2001, 13(2): 54-59. |
| [3] |
俞志明, 陈楠生. 国内外赤潮的发展趋势与研究热点[J]. 海洋与湖沼, 2019, 50(3): 474-486. Yu Zhiming, Chen Nansheng. Emerging trends in red tide and major research progresses[J]. Oceanologia et Limnologia Sinica, 2019, 50(3): 474-486. |
| [4] |
Liu Shuya, Gibson K, Cui Zongmei, et al. Metabarcoding analysis of harmful algal species in Jiaozhou Bay[J]. Harmful Algae, 2020, 92(2): 101722. |
| [5] |
Potter D, Lajeunesse T C, Saunders G W, et al. Convergent evolution masks extensive biodiversity among marine coccoid picoplankton[J]. Biodiversity and Conservation, 1997, 1(6): 99-107. |
| [6] |
Slapeta J, Lopez-Garcia P, Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes[J]. Molecular Biology and Evolution, 2005, 23(1): 23-29. |
| [7] |
Caron D A, Countway P D, Jones A C, et al. Marine protistan diversity[J]. Annual Review of Marine Science, 2012, 4(1): 467-493. |
| [8] |
Rappé M S, Suzuki M T, Vergin K L, et al. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States[J]. Applied and Environmental Microbiology, 1998, 64(1): 294-303. |
| [9] |
Guillou L, Moon-Van Der Staay S Y, Claustre H, et al. Diversity and abundance of bolidophyceae (Heterokonta) in two oceanic regions[J]. Applied and Environmental Microbiology, 1999, 65(10): 4528-4536. |
| [10] |
López-García P, Rodríguez-Valera F, Pedrós-Alió C, et al. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton[J]. Nature, 2001, 409(6820): 603-607. |
| [11] |
Moon-Van Der Staay S Y, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity[J]. Nature, 2001, 409(6820): 607-610. |
| [12] |
Guillou L, Viprey M, Chambouvet A, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata)[J]. Environmental Microbiology, 2008, 10(12): 3349-3365. |
| [13] |
Pawlowski J, Audic S, Adl S, et al. CBOL protist working group:barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms[J]. PLoS Bilogy, 2012, 10(11): e1001419. |
| [14] |
Stern R, Kraberg A, Bresnan E, et al. Molecular analyses of protists in long-term observation programmes-current status and future perspectives[J]. Journal of Plankton Research, 2018, 40(5): 519-536. |
| [15] |
De Vargas C, Audic S, Henry N, et al. Eukaryotic plankton diversity in the sunlit ocean[J]. Science, 2015, 348(6237): 1261605. |
| [16] |
Malviya S, Scalco E, Audic S, et al. Insights into global diatom distribution and diversity in the world's ocean[J]. Proceedings of the National Academy of Sciences, 2016, 113(11): E1516-E1525. |
| [17] |
Le Bescot N, Mahé F, Audic S, et al. Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding[J]. Environmental Microbiology, 2016, 18(2): 609-626. |
| [18] |
Ser-Giacomi E, Zinger L, Malviya S, et al. Ubiquitous abundance distribution of non-dominant plankton across the global ocean[J]. Nature Ecology and Evolution, 2018, 2(8): 1243-1249. |
| [19] |
Grossmann L, Jensen M, Heider D, et al. Protistan community analysis:key findings of a large-scale molecular sampling[J]. The ISME Journal, 2016, 10(9): 2269-2279. |
| [20] |
Logares R, Audic S, Bass D, et al. Patterns of rare and abundant marine microbial eukaryotes[J]. Current Biology, 2014, 24(8): 813-821. |
| [21] |
Piredda R, Claverie J-M, Decelle J, et al. Diatom diversity through HTS-metabarcoding in coastal European seas[J]. Scientific Reports, 2018, 8: 18059. |
| [22] |
Hanson C A, Fuhrman J A, Horner-Devine M C, et al. Beyond biogeographic patterns:processes shaping the microbial landscape[J]. Nature Reviews Microbiology, 2012, 10(7): 497-506. |
| [23] |
Gran-Stadniczeñko S, Egge E, Hostyeva V, et al. Protist diversity and seasonal dynamics in skagerrak plankton communities as revealed by metabarcoding and microscopy[J]. Journal of Eukaryotic Microbiology, 2018, 66(3): 494-513. |
| [24] |
Nagai S, Chen H, Kawakami Y, et al. Monitoring of the toxic dinoflagellate Alexandrium catenella in Osaka Bay, Japan using a massively parallel sequencing (MPS)-based technique[J]. Harmful Algae, 2019, 89: 101660. |
| [25] |
Boulotte N M, Dalton S J, Carroll A G, et al. Exploring the symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals[J]. The ISME Journal, 2016, 10(11): 2693-2701. |
| [26] |
Tragin M, Vaulot D. Green microalgae in marine coastal waters:The Ocean Sampling Day (OSD) dataset[J]. Scientific Reports, 2018, 8: 14020. |
| [27] |
Kopf A, Bicak M, Kottmann R, et al. The ocean sampling day consortium[J]. GigaScience, 2015, 4: 27. |
| [28] |
Goodwin K D, Thompson L R, Duarte B, et al. DNA sequencing as a tool to monitor marine ecological status[J]. Frontiers in Marine Science, 2017, 4: 107. |
| [29] |
Thompson L R, Sanders J G, McDonald D, et al. A communal catalogue reveals Earth's multiscale microbial diversity[J]. Nature, 2017, 551(7681): 457-463. |
| [30] |
陈敏艺, 袁洁, 陈月琴, 等. 海洋超微型浮游植物遗传多样性的分子系统学研究进展[J]. 自然科学进展, 2005, 15(9): 1032-1041. Chen Minyi, Yuan Jie, Chen Yueqin, et al. Advances in molecular phylogeny of the genetic diversity of Marine ultramicroplankton[J]. Progress in Natural Science, 2005, 15(9): 1032-1041. |
| [31] |
林森杰, 王路, 郑连明, 等. 海洋生物DNA条形码研究现状与展望[J]. 海洋学报, 2014, 36(12): 1-17. Lin Senjie, Wang Lu, Zheng Lianming, et al. Current status and future prospect of DNA barcoding in marine biology[J]. Acta Oceanologica Sinica, 2014, 36(12): 1-17. |
| [32] |
陈炼, 吴琳, 刘燕, 等. 环境DNA metabarcoding及其在生态学研究忠的应用[J]. 生态学报, 2016, 36(15): 4573-4582. Chen Lian, Wu Lin, Liu Yan, et al. Application of environmental DNA metabarcoding in ecology[J]. Acta Ecologica Sinica, 2016, 36(15): 4573-4582. |
| [33] |
张宛宛, 谢玉为, 杨江华, 等. DNA宏条形码(metabarcoding)技术在浮游植物群落检测研究中的应用[J]. 生态毒理学报, 2017, 12(1): 15-24. Zhang Wanwan, Xie Yuwei, Yang Jianghua, et al. Applications and prospects of metabarcoding in environmental monitoring of phytoplankton community[J]. Asian Journal of Ecotoxicology, 2017, 12(1): 15-24. |
| [34] |
Xu X, Yu Z, Cheng F, et al. Molecular diversity and ecological characteristics of the eukaryotic phytoplankton community in the coastal waters of the Bohai Sea, China[J]. Harmful Algae, 2017, 61: 13-22. |
| [35] |
宋伦, 吴景, 刘卫东, 等. 渤海长兴岛海域微型和微微型浮游植物多样性[J]. 环境科学研究, 2016, 29(11): 1635-1642. Song Lun, Wu Jing, Liu Weidong, et al. Diversity of marine nanophytoplankton and picophytoplankton in Changxing Island offshore waters of Bohai Sea[J]. Research of Environmental Sciences, 2016, 29(11): 1635-1642. |
| [36] |
宋伦, 吴景, 宋永刚, 等. 褐潮致灾种抑食金球藻在辽东湾的分布[J]. 环境科学研究, 2017, 30(4): 537-544. Song Lun, Wu Jing, Song Yonggang, et al. Distribution of brown tide species aureococcus anophagefferens in Liaodong Bay[J]. Research of Environmental Sciences, 2017, 30(4): 537-544. |
| [37] |
于杰, 张玲玲, 孙妍, 等. 渤海海域褐潮期微型浮游生物多样性的初步研究[J]. 中国海洋大学学报, 2015, 45(3): 73-78. Yu Jie, Zhang Lingling, Sun Yan, et al. Diversity of nanoplankton during the brown tide in the Bohai Sea[J]. Journal of Ocean University of China, 2015, 45(3): 73-78. |
| [38] |
Chen Z, Zhang Q, Kong F, et al. Resolving phytoplankton taxa based on high-throughput sequencing during brown tides in the Bohai Sea, China[J]. Harmful Algae, 2019, 84: 127-138. |
| [39] |
Kong J, Wang Y, Warren A, et al. Diversity distribution and assembly mechanisms of planktonic and benthic microeukaryote communities in intertidal zones of Southeast Fujian, China[J]. Frontiers in Microbiology, 2019, 10: 2640. |
| [40] |
Shih C Y, Lu H M, Gong G C, et al. High diversity of haptophytes in the East China Sea revealed by next-generation sequencing and scanning electron microscopy[J]. Journal of Oceanography, 2019, 75(4): 305-317. |
| [41] |
Qin Z, Yu K, Chen B, et al. Diversity of Symbiodiniaceae in 15 coral species from the southern South China Sea:potential relationship with coral thermal adaptability[J]. Frontiers in Microbiology, 2019, 10: 2343. |
| [42] |
Chen B, Yu K, Liang J, et al. Latitudinal variation in the molecular diversity and community composition of Symbiodiniaceae in coral from the South China Sea[J]. Frontiers in Microbiology, 2019, 10: 1278. |
| [43] |
Gong S, Chai G, Xiao Y, et al. Flexible symbiotic associations of symbiodinium with five typical coral species in tropical and subtropical reef regions of the northern South China Sea[J]. Frontiers in Microbiology, 2018, 9: 2485. |
| [44] |
刘永健, 杨官品, 管晓菁. 胶州湾浮游植物rbcL基因分子遗传多样性研究[J]. 应用生态学报, 2004, 15(9): 1626-1632. Liu Yongjian, Yang Guanpin, Guan Xiaojing. Molecular genetic diversity of phytoplankton rbcL gene in Jiaozhou Bay[J]. Chinese Journal of Applied Ecology, 2004, 15(9): 1626-1632. |
| [45] |
Wu W, Logares R, Huang B, et al. Abundant and rare picoeukaryotic sub-communities present contrasting patterns in the epipelagic waters of marginal seas in the northwestern Pacific Ocean[J]. Environmental Microbiology, 2017, 19(1): 287-300. |
| [46] |
Jiang Z Y, Wang Y S, Cheng H, et al. Variation of phytoplankton community structure from the Pearl River estuary to South China Sea[J]. Ecotoxicology, 2015, 24(7-8): 1442-1449. |
| [47] |
Treusch A H, Demir-Hilton E, Vergin K L, et al. Phytoplankton distribution patterns in the northwestern Sargasso Sea revealed by small subunit rRNA genes from plastids[J]. ISME Journal, 2012, 6(3): 481-492. |
| [48] |
Liu H, Probert I, Uitz J, et al. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(31): 12803-12808. |
| [49] |
Callahan B J, Wong J, Heiner C, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution[J]. Nucleic Acids Research, 2019, 47(18): e103. |
| [50] |
Callahan B J, McMurdie P J, Holmes S P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis[J]. The ISME Journal, 2017, 11(12): 2639-2643. |
| [51] |
Callahan B J, McMurdie P J, Rosen M J, et al. DADA2:High-resolution sample inference from Illumina amplicon data[J]. Nature Methods, 2016, 13(7): 581-583. |
| [52] |
Rosen M J, Callahan B J, Fisher D S, et al. Denoising PCR-amplified metagenome data[J]. BMC Bioinformatics, 2012, 13: 283. |
| [53] |
Knight R, Vrbanac A, Taylor B C, et al. Best practices for analysing microbiomes[J]. Nature Reviews Microbiology, 2018, 16(7): 410-422. |
| [54] |
Amir A, McDonald D, Navas-Molina J A, et al. Deblur rapidly resolves single-nucleotide community sequence patterns[J]. mSystems, 2017, 2(2): e00191-16. |
| [55] |
Quince C, Lanzen A, Davenport R J, et al. Removing noise from pyrosequenced amplicons[J]. BMC Bioinformatics, 2011, 12: 1-18. |
| [56] |
林凤翱, 卢兴旺, 洛昊, 等. 渤海赤潮的历史、现状及其特点[J]. 海洋环境科学, 2008, 27(增刊2): 1-5. |
| [57] |
宋南奇, 王诺, 吴暖, 等. 基于GIS的我国渤海1952~2016年赤潮时空分布[J]. 中国环境科学, 2018, 38(3): 1142-1148. Song Nanqi, Wang Nuo, Wu Nuan, et al. Temporal and spatial distribution of harmful algal blooms in the Bohai Sea during 1952~2016 based on GIS[J]. China Environmental Science, 2018, 38(3): 1142-1148. |
| [58] |
喻龙, 郝彦菊. 烟台四十里湾一次血红哈卡藻赤潮过程的分析[J]. 海洋科学进展, 2009, 27(4): 516-522. Yu Long, Hao Yanju. Process analysis for harmful bloom of Akashiwo sanguinea in Sishili Bay of Yantai[J]. Advances in Marine Science, 2009, 27(4): 516-522. |
| [59] |
尹翠玲, 张秋丰, 刘洋, 等. 2012年渤海湾赤潮监控区赤潮发生及富营养化状况[J]. 海洋湖沼通报, 2014(1): 137-142. Yin Cuiling, Zhang Qiufeng, Liu Yang, et al. Red tide occurrence and eutrofication status in the red tide monitoring area of Bohai Bay in 2012[J]. Transactions of Oceanology and Limnology, 2014(1): 137-142. |
| [60] |
孙雪梅, 徐东会, 夏斌, 等. 渤海中部网采浮游植物种类组成和季节变化[J]. 渔业科学进展, 2016, 37(4): 19-27. Sun Xuemei, Xu Donghui, Xia Bin, et al. Species composition and seasonal variation of netz-phytoplankton in the central Bohai Sea[J]. Progress in Fishery Sciences, 2016, 37(4): 19-27. |
| [61] |
Yao P, Lei L, Zhao B, et al. Spatial-temporal variation of Aureococcus anophagefferens blooms in relation to environmental factors in the coastal waters of Qinhuangdao, China[J]. Harmful Algae, 2019, 86: 106-118. |
| [62] |
宋伦, 刘卫东, 吴景, 等. 有害甲藻Stoeckeria algicida在辽东湾的时空分布[J]. 生态学报, 2017, 37(4): 1339-1345. Song Lun, Liu Weidong, Wu Jing, et al. Distribution of the toxic dinoflagellate Stoeckeria algicida in Liaodong Bay[J]. Acta Ecologica Sinica, 2017, 37(4): 1339-1345. |
| [63] |
Jeong H J, Kim J S, Park J Y, et al. Stoeckeria algicida n. gen., n. sp. (Dinophyceae) from the coastal waters off Southern Korea:morphology and small subunit ribosomal DNA gene sequence[J]. Journal of Eukaryotic Microbiology, 2005, 52(4): 382-390. |
| [64] |
Zhang Q C, Qiu L M, Yu R C, et al. Emergence of brown tides caused by Aureococcus anophagefferens Hargraves et Sieburth in China[J]. Harmful Algae, 2012, 19: 117-124. |
| [65] |
Zhen Y, Qiao L, Gu B, et al. Characteristics of eukaryotic microalgal community and its abiotic influencing factors during brown tide blooms near Qinhuangdao, China[J]. Harmful Algae, 2016, 57: 1-12. |
| [66] |
Tang Y Z, Ma Z, Hu Z, et al. 3, 000 km and 1, 500-year presence of Aureococcus anophagefferens reveals indigenous origin of brown tides in China[J]. Molecular Ecology, 2019, 28(17): 4065-4076. |
| [67] |
Qiao L, Yu J, Li Y, et al. Succession of the Nanoplankton Community during a Brown Tide in a Scallop Culture Area in China[J]. Ocean Science Journal, 2019, 54(3): 375-392. |
| [68] |
Song L, Wu J, Du J, et al. The Characteristics and distribution of eukaryotic phytoplankton community in Liaodong Bay, China[J]. Ocean Science Journal, 2019, 54(2): 183-203. |
| [69] |
Song L, Wu J, Du J, et al. Comparison of two methods to assess the size structure of phytoplankton community assemblages, in Liaodong Bay, China[J]. Journal of Ocean University of China, 2019, 18(5): 1207-1215. |
| [70] |
Song H, Ji R, Xin M, et al. Spatial heterogeneity of seasonal phytoplankton blooms in a marginal sea:physical drivers and biological responses[J]. ICES Journal of Marine Science, 2020, 77(1): 408-418. |
| [71] |
Huo Y, Shi H, Zhang J, et al. Spatio-temporal variability of phytoplankton assemblages and its controlling factors in spring and summer in the Subei Shoal of Yellow Sea, China[J]. Acta Oceanologica Sinica, 2019, 38(10): 84-92. |
| [72] |
石雅君, 刘东艳, 邵红兵, 等. 北黄海表层沉积物中甲藻孢囊的分布特征[J]. 海洋通报, 2011, 30(3): 320-327. Shi Yajun, Liu Dongyan, Shao Hongbing, et al. Distribution of dinoflagellate cysts in the surface sediments from the northern Yellow Sea, China[J]. Marine Science Bulletin, 2011, 30(3): 320-327. |
| [73] |
Liu P, Wang W, Luo Q, et al. Characteristic of eukaryotic plankton community and its correlation with environmental factors in the northern seas of Shandong Peninsula[J]. Oceanologia et Liminologia Sinica, 2019, 50(4): 876-885. |
| [74] |
Xu Q, Sukigara C, Goes J I, et al. Interannual changes in summer phytoplankton community composition in relation to water mass variability in the East China Sea[J]. Journal of Oceanography, 2019, 75(1): 61-79. |
| [75] |
Zhao Y, Yu R C, Kong F Z, et al. Distribution patterns of picosized and nanosized phytoplankton assemblages in the East China Sea and the Yellow Sea:implications on the impacts of Kuroshio intrusion[J]. Journal of Geophysical Research:Oceans, 2019, 124(2): 1262-1276. |
| [76] |
Wang J, Wu J. Occurrence and potential risks of harmful algal blooms in the East China Sea[J]. Science of the Total Environment, 2009, 407(13): 4012-4021. |
| [77] |
Zhou Z X, Yu R C, Sun C, et al. Impacts of Changjiang River discharge and Kuroshio intrusion on the diatom and dinoflagellate blooms in the East China Sea[J]. Journal of Geophysical Research:Oceans, 2019, 124(7): 5244-5257. |
| [78] |
周茜茜, 陈长平, 梁君荣, 等. 中国东部陆架边缘海网采浮游植物种类组成和季节变化[J]. 生物多样性, 2015, 23(1): 23-32. Zhou Qianqian, Chen Changping, Liang Junrong, et al. Species composition and seasonal variation of netz-phytoplankton in the eastern marginal China seas[J]. Biodiversity Science, 2015, 23(1): 23-32. |
| [79] |
Guo S, Feng Y, Wang L, et al. Seasonal variation in the phytoplankton community of a continental-shelf sea:the East China Sea[J]. Marine Ecology progress Series, 2014, 516: 103-126. |
| [80] |
Lu D D, Goebel J. Five red tide species in genus Prorocentrum including the description of Prorocentrum donghaiense Lu sp. nov. from the East China Sea[J]. Chinese Journal of Oceanology and Limnology, 2001, 19(4): 337-344. |
| [81] |
Shin H H, Li Z, Seo M H, et al. Harmful dinoflagellate Prorocentrum donghaiense Lu is widely distributed along the East China Sea and Korean coastal area[J]. Ocean Science Journal, 2019, 54(4): 685-691. |
| [82] |
Lu D, Goebel J, Qi Y, et al. Morphological and genetic study of Prorocentrum donghaiense Lu from the East China Sea, and comparison with some related Prorocentrum species[J]. Harmful Algae, 2005, 4(3): 493-505. |
| [83] |
陆斗定, 齐雨藻, GoebelJ, 等. 东海原甲藻修订及与相关原甲藻的分类学比较[J]. 应用生态学报, 2003, 14(7): 1060-1064. Lu Douding, Qi Yuzao, Goebel J, et al. Redescription of Prorocentrum donghaiense Lu and comparison with relevant Prorocentrum species[J]. Chinese Journal of Applied Ecology, 2003, 14(7): 1060-1064. |
| [84] |
Cen J, Wang J, Huang L, et al. Who is the "murderer" of the bloom in coastal waters of Fujian, China, in 2019?[J]. Journal of Oceanology and Limnology, 2019, 38(3): 722-732. |
| [85] |
Luo Z, Yang W, Leaw C P, et al. Cryptic diversity within the harmful dinoflagellate Akashiwo sanguinea in coastal Chinese waters is related to differentiated ecological niches[J]. Harmful Algae, 2017, 66: 88-96. |
| [86] |
Aifeng L, Rencheng Y, Yunfeng W, et al. Morphological and toxicity characteristics of Pseudo-nitzschia pungens strain PP0201-01 isolated from the East China Sea[J]. Chinese Journal of Oceanology and Limnology, 2005, 23(4): 418-426. |
| [87] |
Chen W, Pan Y, Yu L, et al. Patterns and processes in marine microeukaryotic community biogeography from Xiamen coastal waters and intertidal sediments, southeast China[J]. Frontiers in Microbiology, 2017, 8: 1912. |
| [88] |
Yu L, Zhang W, Liu L, et al. Determining microeukaryotic plankton community around Xiamen island, southeast China, using Illumina MiSeq and PCR-DGGE techniques[J]. PLoS ONE, 2015, 10(5): e0127721. |
| [89] |
Kang L K, Lu H M, Sung P T, et al. The summer distribution of coccolithophores and its relationship to water masses in the East China Sea[J]. Journal of Oceanography, 2016, 72(6): 883-893. |
| [90] |
Sun J, Gu X Y, Feng Y Y, et al. Summer and winter living coccolithophores in the Yellow Sea and the East China Sea[J]. Biogeosciences, 2014, 11(3): 779-806. |
| [91] |
Yang T N. Summer and winter distribution and malformation of coccolithophores in the East China Sea[J]. Micropaleontology, 2004, 50(Suppl 1): 157-170. |
| [92] |
李红飞, 林森杰. 南海浮游植物生态学研究进展[J]. 厦门大学学报(自然科学版), 2019, 58(1): 1-10. Li Hongfei, Lin Senjie. Research progresses of phytoplankton ecology in the South China Sea[J]. Journal of Xiamen University(Natural Science), 2019, 58(1): 1-10. |
| [93] |
Wei N, Satheeswaran T, Jenkinson I R, et al. Factors driving the spatiotemporal variability in phytoplankton in the Northern South China Sea[J]. Continental Shelf Research, 2018, 162: 48-55. |
| [94] |
朱根海, 宁修仁, 蔡昱明, 等. 南海浮游植物种类组成和丰度分布的研究[J]. 海洋学报, 2003, 25(增刊2): 8-23. Zhu Genhai, Ning Xiuren, Cai Yuming, et al. Studies on species composition and abundance distribution of phytoplankton in the South China Sea[J]. Acta Oceanologica Sinica, 2003, 25(supplement 2): 8-23. |
| [95] |
孙军, 宋书群, 乐凤凤, 等. 2004年冬季南海北部浮游植物[J]. 海洋学报, 2007, 29(5): 132-145. Sun Jun, Song Shuqun, Le Fengfeng, et al. Phytoplankton in northern South China Sea in the winter of 2004[J]. Acta Oceanologica Sinica, 2007, 29(5): 132-145. |
| [96] |
马威, 孙军. 夏、冬季南海北部浮游植物群落特征[J]. 生态学报, 2014, 34(3): 621-632. Ma Wei, Sun Jun. Characteristics of phytoplankton community in the northern South China Sea in summer and winter[J]. Acta Ecologica Sinica, 2014, 34(3): 621-632. |
| [97] |
马威, 孙军, 薛冰, 等. 2010年秋季南海北部浮游植物群落结构研究[J]. 海洋学报, 2016, 38(4): 43-53. Ma Wei, Sun Jun, Xue Bing, et al. Phytoplankton Community Structure of northern South China Sea in autumn of 2010[J]. Acta Oceanologica Sinica, 2016, 38(4): 43-53. |
| [98] |
魏玉秋, 孙军, 丁昌玲. 2014年夏季南海北部超微型浮游植物分布及环境因子影响[J]. 海洋学报, 2015, 37(12): 56-65. Wei Yuqiu, Sun Jun, Ding Changling. Distribution and environmental impact factors of picoplankton in the northern South China Sea in summer 2014[J]. Acta Oceanologica Sinica, 2015, 37(12): 56-65. |
| [99] |
Li L, Lü S, Cen J. Spatio-temporal variations of Harmful algal blooms along the coast of Guangdong, Southern China during 1980-2016[J]. Journal of Oceanology and Limnology, 2019, 37(2): 535-551. |
| [100] |
Liu H X, Huang H H, Xu S N, et al. Planktonic community structure during a harmful bloom of Phaeocystis globosa in a subtropical bay, with special reference to the ciliate assemblages[J]. Ecotoxicology, 2015, 24(7-8): 1419-1429. |
| [101] |
黄长江, 董巧香, 郑磊. 1997年底中国东南沿海大规模赤潮原因生物的形态分类与生态学特征[J]. 海洋与湖沼, 1999, 30(6): 581-590. Huang Changjiang, Dong Qiaoxiang, Zheng Lei. Taxonomic andeoclogical studies on a large scale Phaeocystis pouchetii bloom in the southeast coast of China during late 1997[J]. Oceanologia et Limnologia Sinica, 1999, 30(6): 581-590. |
| [102] |
袁洁, 邵鹏, 陈月琴, 等. 南沙群岛微型与超微型真核藻类遗传多样性的初步研究[J]. 海洋科学, 2003, 27(7): 43-47. Yuan Jie, Shao Peng, Chen Yueqin, et al. Primary study of genetic diversity of eukaryotic micro- and pico- plankton in Nansha Islands[J]. Marine Sciences, 2003, 27(7): 43-47. |
| [103] |
Shen P P, Tan Y H, Huang L M, et al. Occurrence of brackish water phytoplankton species at a closed coral reef in Nansha Islands, South China Sea[J]. Marine Pollution Bulletin, 2010, 60(10): 1718-1725. |
| [104] |
Liu H, Song X, Huang L, et al. Diurnal variation of phytoplankton community in a high frequency area of HABs:Daya Bay, China[J]. Chinese Journal of Oceanology and Limnology, 2011, 29(4): 800-806. |
| [105] |
Wang J J, Tang D L. Phytoplankton patchiness during spring intermonsoon in western coast of South China Sea[J]. Deep-sea Research Part Ⅱ-Topical Studies in Oceanography, 2014, 101(SI): 120-128. |
| [106] |
Tang D, Kawamura H, Van Dien T, et al. Offshore phytoplankton biomass increase and its oceanographic causes in the South China Sea[J]. Marine Ecology Progress Series, 2004, 268: 31-41. |
| [107] |
Tang D L, Kawamura H, Doan-Nhu H, et al. Remote sensing oceanography of a harmful algal bloom off the coast of southeastern Vietnam[J]. Journal of Geophysical Research-Oceans, 2004, 109: C03014. |
| [108] |
Tang D L, Kawamura H, Shi P, et al. Seasonal phytoplankton blooms associated with monsoonal influences and coastal environments in the sea areas either side of the Indochina Peninsula[J]. Journal of Geophysical Research-Biogeosciencs, 2006, 111: G01010. |
| [109] |
Hai D N, Lam N N, Dippner J W. Development of Phaeocystis globosa blooms in the upwelling waters of the South Central coast of Viet Nam[J]. Journal of Marine systems, 2010, 83(3-4): 253-261. |
| [110] |
Cheung M K, Au C H, Chu K H, et al. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing[J]. ISME JOURNAL, 2010, 4(8): 1053-1059. |
| [111] |
Li L, Huang Q, Wu S, et al. The spatial and temporal distribution of microalgae in the South China Sea:evidence from GIS-based analysis of 18S rDNA sequences[J]. Science in China Series C-Life Sciences, 2008, 51(12): 1121-1128. |
| [112] |
杨世民, 王丽莎, 石晓勇. 2009年春季胶州湾浮游植物群落结构特征[J]. 海洋与湖沼, 2014, 45(6): 1234-1240. Yang Shimin, Wang Lisha, Shi Xiaoyong. Phytoplankton community of the Jiaozhou Bay in Spring 2009[J]. Oceanologia et Limnologia Sinica, 2014, 45(6): 1234-1240. |
| [113] |
Guo S, Zhu M, Zhao Z, et al. Spatial-temporal variation of phytoplankton community structure in Jiaozhou Bay, China[J]. Journal of Oceanology and Limnology, 2019, 37(5): 1611-1624. |
| [114] |
Shen Z L. Historical changes in nutrient structure and its influences on phytoplantkon composition in Jiaozhou Bay[J]. Estuarine, Coastal and Shelf Science, 2001, 52(2): 211-224. |
| [115] |
杨广普, 江涛, 赵永芳, 等. 基于长时序遥感影像的胶州湾秋季叶绿素a浓度变化及其影响因素研究[J]. 海洋学报, 2019, 41(1): 183-190. Yang Guangpu, Jiang Tao, Zhao Yongfang, et al. Study on variation in chlorophyⅡ a concentration and its influencing factors of Jiaozhou Bay in autumn based on long term remote sensing images[J]. Acta Oceanologica Sciences, 2019, 41(1): 183-190. |
| [116] |
李影, 汤亚楠, 沈萍萍, 等. 胶州湾表层沉积物中甲藻孢囊的分布[J]. 海洋与湖沼, 2017, 48(4): 760-766. Li Ying, Tang Yanan, Shen Pingping, et al. Distribution of dinoflagellate resting cysts in surface sediment of Jiaozhou Bay, China[J]. Oceanologia et Limnologia Sinica, 2017, 48(4): 760-766. |
| [117] |
Sun X, Sun S. Conduct long-term observations of coastal ecosystems and lead the marine ecosystem health research[J]. Bulletin of the Chinese Academy of Sciences, 2019, 34(12): 1458-1466. |
| [118] |
韩笑天, 邹景忠, 张永山. 胶州湾赤潮生物种类及其生态分布特征[J]. 海洋科学, 2004, 28(2): 49-54. Han Xiaotian, Zou Jingzhong, Zhang Yongshan. Harmful algae bloom species in Jiaozhou Bay and the features of distribution[J]. Marine Sciences, 2004, 28(2): 49-54. |
| [119] |
田达玮, 宋书群, 陈田田, 等. 胶州湾夜光藻种群动态及其大量繁殖的生态机制[J]. 海洋与湖沼, 2017, 48(2): 276-284. Tian Dawei, Song Shuqun, Chen Tiantian, et al. Noctiluca sccintillans blooming in the Jiaozhou Bay:population dynamics and ecological mechanism[J]. Oceanologia et Limnologia Sinica, 2017, 48(2): 276-284. |
| [120] |
Wang W, Sun S, Sun X, et al. Seasonal phenology of the heterotrophic dinoflagellate Noctiluc ascintillans (Macartney) in Jiaozhou Bay and adjacent coastal Yellow Sea, China[J]. Journal of Oceanology and Limnology, 2018, 36(4): 1280-1293. |
| [121] |
卢敏, 张龙军, 李超, 等. 1999年7月胶州湾东部赤潮生消过程生态环境要素分析[J]. 黄渤海海洋, 2001, 19(4): 43-50. Lu Min, Zhang Longjun, Li Chao, et al. Analysis of the ecological environment elements in the red tide generating and vanishing process in the eastern Jiaozhou Bay in July, 1999[J]. Yellow and Bohai Seas, 2001, 19(4): 43-50. |
| [122] |
张永山, 吴玉霖, 邹景忠, 等. 胶州湾浮动弯角藻赤潮生消过程[J]. 海洋与湖沼, 2002, 33(1): 55-61. Zhang Yongshan, Wu Yulin, Zou Jingzhong, et al. A red tide caused by diatom Eucampia zoodiacus in the Jiaozhou Bay[J]. Oceanologia et Limnologia Sinica, 2002, 33(1): 55-61. |
| [123] |
霍文毅, 俞志明, 邹景忠, 等. 胶州湾浮动弯角藻赤潮生消动态过程及其成因分析[J]. 水产学报, 2001, 25(3): 222-226. Huo Wenyi, Yu Zhiming, Zou Jingzhong, et al. Analysis of dynamic process and the causes of Eucampia zoodiacus red tide in Jiaozhou Bay[J]. Journal of Fisheries of China, 2001, 25(3): 222-226. |
| [124] |
霍文毅, 俞志明, 邹景忠, 等. 胶州湾中肋骨条藻赤潮与环境因子的关系[J]. 海洋与湖沼, 2001, 32(3): 311-318. Huo Wenyi, Yu Zhiming, Zou Jingzhong, et al. Outbreak of Skeletonema costatum red tide and its relations to environmental factors in Jiaozhou Bay[J]. Oceanologia et Limnologia Sinica, 2001, 32(3): 311-318. |
| [125] |
殷明焱, 胡晓燕, 曾呈奎. 胶州湾有毒藻华形成种里氏金色藻(Chrysochromulina leadbeateri)的分类学研究[J]. 海洋与湖沼, 2009, 40(6): 799-802. Yin Mingyan, Hu Xiaoyan, Zeng Chengkui. Taxonomy of Chrysochromulina leadbeateri (Prymnesiophyceae), a toxic bloom-forming species iolated from Jiaozhou Bay, China[J]. Oceanologia et Limnologia Sinica, 2009, 40(6): 799-802. |
| [126] |
Xu Y, Zhang T, Zhou J. Historical occurrence of algal blooms in the northern Beibu Gulf of China and implications for future trends[J]. Frontiers in Microbiology, 2019, 10: 451. |
| [127] |
Kaiser D, Unger D, Qiu G, et al. Natural and human influences on nutrient transport through a small subtropical Chinese estuary[J]. Science of the Total Environment, 2013, 450: 92-107. |
| [128] |
沈萍萍, 齐雨藻, 欧林坚. 中国沿海球形棕囊藻(Phaeocystis globosa)的分类、分布及其藻华[J]. 海洋科学, 2018, 42(10): 146-162. Shen Pingping, Qi Yuzao, Ou Linjian. Phaeocystis globosa in coastal China:taxonomy, distribution, and its blooms[J]. Marine Sciences, 2018, 42(10): 146-162. |
| [129] |
刘国强, 史海燕, 魏春雷, 等. 广西涠洲岛海域浮游植物和赤潮生物种类组成的初步研究[J]. 海洋通报, 2008, 27(3): 43-48. Liu Guoqiang, Shi Haiyan, Wei Chunlei, et al. Preliminary studies on phytoplankton and red tide organism composition in the coastal water of the Weizhou Island, Guangxi[J]. Marine Science Bulletin, 2008, 27(3): 43-48. |
| [130] |
胡章喜, 邓蕴彦, 唐赢中. 我国北部湾球形棕囊藻(Phaeocystis globosa)的表面形态和细胞超微结构的电镜观察[J]. 海洋与湖沼, 2019, 50(3): 621-629. Hu Zhangxi, Deng Yunyan, Tang Yingzhong. Scanning and transmission electron microscopy observation on morphology and ultrastructure of Phaeocystis globosa from Beibu Gulf, China[J]. Oceanologia et Limnologia Sinica, 2019, 50(3): 621-629. |
| [131] |
赵越, 于仁成, 张清春, 等. 北部湾海域微型、微微型浮游植物类群季节变化及其与棕囊藻赤潮的关系初探[J]. 海洋与湖沼, 2019, 50(3): 590-600. Zhao Yue, Yu Rencheng, Zhang Qingchun, et al. Relationship between seasonal variation of pico- and nano-phytoplankton assemblages and Phaeocystis red tides in Beibu Gulf[J]. Oceanologia et Limnologia Sinica, 2019, 50(3): 590-600. |
| [132] |
姜发军, 陈波, 何碧娟, 等. 广西钦州湾浮游植物群落结构特征[J]. 广西科学, 2012, 19(3): 268-275. Jiang Fajun, Chen Bo, He Bijuan, et al. Phytoplankton community in coastal waters of Qinzhou Bay in Guangxi[J]. Guangxi Sciences, 2012, 19(3): 268-275. |
| [133] |
贺成, 宋书群, 李才文. 广西北部湾海域球形棕囊藻囊体时空分布及其影响因素[J]. 海洋与湖沼, 2019, 50(3): 630-643. He Cheng, Song Shuqun, Li Caiwen. The spatial-temperal distribution of Phaeocystis globosa colonies and related affecting factors in Guangxi Beibu Guf[J]. Oceanologia et Limnologia Sinica, 2019, 50(3): 630-643. |
| [134] |
Gong B, Wu H, Ma J, et al. The algae community in taxon Haptophyceae at the early bloom stage of Phaeocystis globosa in Northern Beibu Gulf in winter[J]. bioRxiv, 2018, http://dx.doi.org/10.1101/492454.
|
| [135] |
周名江, 颜天, 邹景忠. 长江口邻近海域赤潮发生区基本特征初探[J]. 应用生态学报, 2003, 14(7): 1031-1038. Zhou Mingjiang, Yan tian, Zou Jingzhong. Preliminary analysis of the characteristics of red tide areas in Changjiang River estuary and its adjacent sea[J]. Chinese Journal of Applied Ecology, 2003, 14(7): 1031-1038. |
| [136] |
Zhou Z, Yu R C, Zhou M. Resolving the complex relationship between harmful algal blooms and environmental factors in the coastal waters adjacent to the Changjiang River estuary[J]. Harmful Algae, 2017, 62: 60-72. |
| [137] |
Jiang Z, Liu J, Chen J, et al. Responses of summer phytoplankton community to drastic environmental changes in the Changjiang (Yangtze River) estuary during the past 50 years[J]. Water Research, 2014, 54: 1-11. |
| [138] |
于仁成, 张清春, 孔凡洲, 等. 长江口及其邻近海域有害藻华的发生情况、危害效应与演变趋势[J]. 海洋与湖沼, 2017, 48(6): 1178-1186. Yu Rencheng, Zhang Qingchun, Kong Fanzhou, et al. Status, impacts and long-term changes of harmful algal blooms in the sea area adjacent to the Changjiang River Estuary[J]. Oceanologia et Limnologia Sinica, 2017, 48(6): 1178-1186. |
| [139] |
Wang B. Cultural eutrophication in the Changjiang (Yangtze River) plume:history and perspective[J]. Estuarine Coastal and Shelf Science, 2006, 69(3-4): 471-477. |
| [140] |
王金辉. 长江口邻近水域的赤潮生物[J]. 海洋环境科学, 2002, 21(2): 37-41. Wang Jinhui. HAB algae nearby Changjiang Estuary[J]. Marine Environmental Science, 2002, 21(2): 37-41. |
| [141] |
洪君超, 黄秀清, 蒋晓山, 等. 长江口中肋骨条藻赤潮发生过程环境要素分析-营养盐状况[J]. 海洋与湖沼, 1994, 25(2): 179-184. Honh Junchao, Huang Xiuqing, Jiang Xiaoshan, et al. Principle environmental facor during red tide outbreak of Skeletonema costatum in Yangtse Estuary: nutrients status[J]. Oceanologia et Limnologia Sinica, 1994, 25(2): 179-184. |
| [142] |
黄秀清, 蒋晓山, 王桂兰, 等. 长江口中肋骨条藻赤潮发生过程环境要素分析:水温、盐度、DO和pH特征[J]. 海洋通报, 1994, 13(4): 35-40. Huang Xiuqing, Jiang Xiaoshan, Wang Guilan, et al. Principle environmental factors during red tide outbreak of Skeletonema costatum in Yangtse Estuary: water temperature, salinity, DO and pH[J]. Marine Science Bulletin, 1994, 13(4): 35-40. |
| [143] |
沈竑, 洪君超. 长江口中肋骨条藻赤潮发生全过程调查报告-浮游植物群落结构及细胞形态研究[J]. 海洋与湖沼, 1994, 25(6): 591-595. Shen Hong, Hong Junchao. Investigation report on the Skeletonema costatum red tide in Changjiang River Estuary-study on the phytoplankton community composition and cell morphology[J]. Oceanologia et Limnologia Sinica, 1994, 25(6): 591-595. |
| [144] |
吴玉霖, 傅月娜, 张永山, 等. 长江口海域浮游植物分布及其与径流的关系[J]. 海洋与湖沼, 2004, 35(3): 246-251. Wu Yulin, Fu Yuena, Zhang Yongshan, et al. Phytoplankton distribution and its relation to the runoff in the Changjiang (Yangzte) Estuary[J]. Oceanologia et Limnologia Sinica, 2004, 35(3): 246-251. |
| [145] |
Gao X, Song J. Phytoplankton distributions and their relationship with the environment in the Changjiang Estuary, China[J]. Marine Pollution Bulletin, 2005, 50(3): 327-335. |
| [146] |
栾青杉, 孙军. 2005年夏季长江口水域浮游植物群集特征及其与环境因子的关系[J]. 生态学报, 2010, 30(18): 4967-4975. Luan Qingshan, Sun Jun. Feature of phytoplankton assemblages in Yangtze River estuary and its relationship with environmental factors in summer 2005[J]. Acta Ecologica Sinica, 2010, 30(18): 4967-4975. |
| [147] |
朱根海, 许卫忆, 朱德第, 等. 长江口赤潮高发区浮游植物与水动力环境因子的分布特征[J]. 应用生态学报, 2003, 14(7): 1135-1139. Zhu Genhai, Xu Weiyi, Zhu Dedi, et al. Distribution of phytoplankton and water dynamical environmental factors in high red tide occurrence area of Changjiang River estuary[J]. Chinese Journal of Applied Ecology, 2003, 14(7): 1135-1139. |
| [148] |
王丹, 孙军, 周锋, 等. 2006年6月长江口低氧区及邻近水域浮游植物[J]. 海洋与湖沼, 2008, 39(6): 619-627. Wang Dan, Sun Jun, Zhou Feng, et al. Phytoplanton of Changjiang (Yangtze River) Estuary hypoxia are and the adjacent east China Sea in June, 2006[J]. Oceanologia et Limnologia Sinica, 2008, 39(6): 619-627. |
| [149] |
唐峰华, 伍玉梅, 樊伟, 等. 长江口浮游植物分布情况及与径流关系的初步探讨[J]. 生态环境学报, 2010, 19(12): 2934-2940. Tang Fenghua, Wu Yumei, Fan Wei, et al. Preliminary discussion on phytoplankton distribution and its relation to the runoff in the Yangtze River Estuary[J]. Ecology and Environment, 2010, 19(12): 2934-2940. |
| [150] |
徐兆礼, 白雪梅, 袁骐, 等. 长江口浮游植物生态研究[J]. 中国水产学报, 1999, 6(5): 52-54. Xu Zhaoli, Bai Xuemei, Yuan Qi, et al. An ecological study on phytoplankton in the Changjiang Estuary[J]. Journal of Fisheries of China, 1999, 6(5): 52-54. |
| [151] |
王云龙, 袁骐, 沈新强. 长江口及邻近水域春季浮游植物的生态特征[J]. 中国水产科学, 2005, 12(3): 300-306. Wang Yunlong, Yuan Qi, Shen Xinqiang. Ecological character of phytoplankton in spring in the Yangtze River estuary and adjacent waters[J]. Journal of Fishery Sciences of China, 2005, 12(3): 300-306. |
| [152] |
顾新根, 袁骐, 杨蕉文, 等. 长江口羽状锋海区浮游植物的生态研究[J]. 中国水产科学, 1995, 2(1): 1-15. Gu Xingen, Yuan Qi, Yang Jiaowen, et al. An ecological study on phytoplankton in frontal region of Changjiang Estuarine area[J]. Journal of Fishery Sciences of China, 1995, 2(1): 1-15. |
| [153] |
顾新根, 袁骐, 沈焕庭, 等. 长江口最大浑浊带浮游植物的生态研究[J]. 中国水产科学, 1995, 2(1): 16-27. Gu Xingen, Yuan Qi, Shen Huanting, et al. The ecological study of phytoplankton in maximun turbid zone of Changjiang estuary[J]. Journal of Fishery Sciences of China, 1995, 2(1): 16-27. |
| [154] |
杨蕉文, 华棣, 顾新根. 长江口羽状锋海区浮游植物的生态研究-昼夜分布动态[J]. 东海海洋, 1994, 12(1): 47-57. Yang Jiaowen, Hua Di, Gu Xingen. An ecological study of phytoplankton in the pinklike frontage of the Yangtze estuary-diurnal distribution dynamics[J]. Donghai Marine Science, 1994, 12(1): 47-57. |
| [155] |
林梅, 李杨. 长江口海域春季浮游植物的年际变化[J]. 海洋与湖沼, 2017, 48(2): 303-311. Lin Mei, Li Yang. Interdecadal variation of phytoplankton assemblage in Changjiang River Estuary in spring[J]. Oceanologia et Limnologia Sinica, 2017, 48(2): 303-311. |
| [156] |
王金辉, 黄秀清, 刘阿成, 等. 长江口及邻近水域的生物多样性变化趋势分析[J]. 海洋通报, 2004, 23(1): 32-39. Wang Jinhui, Huang Xiuqing, Liu A'cheng, et al. Tendency of the biodiversity variation nearby Changjiang Estuary[J]. Marine Science Bulletin, 2004, 23(1): 32-39. |
| [157] |
王金辉. 长江口3个不同生态系的浮游植物群落[J]. 中国海洋大学学报, 2002, 32(3): 422-428. Wang Jinhui. Phytoplankton communities in three distinct ecotypes of the Changjiang Estuary[J]. Journal of Ocean University of China, 2002, 32(3): 422-428. |
| [158] |
田伟, 孙军, 樊孝鹏, 等. 2008年春季东海近海浮游植物群落[J]. 海洋科学进展, 2010, 28(2): 170-178. Tian Wei, Sun Jun, Fan Xiaopeng, et al. Phytoplankton community in coastal waters of the East China Sea in spring 2008[J]. Advances in Marine Science, 2010, 28(2): 170-178. |
| [159] |
高亚辉, 虞秋波, 齐雨藻, 等. 长江口附近海域春季浮游硅藻的种类组成和生态分布[J]. 应用生态学报, 2003, 14(7): 1044-1048. Gao Yahui, Yu Qiubo, Qi Yuzao, et al. Species composition and ecological distribution of planktonic diatoms in the Changjiang River estuary during Spring[J]. Chinese Journal of Applied Ecology, 2003, 14(7): 1044-1048. |
| [160] |
李照, 宋书群, 李才文, 等. 丰、枯水期长江口邻近海域浮游植物群落结构特征及其环境影响初探[J]. 海洋学报, 2017, 39(10): 124-144. Li Zhao, Song Shuqun, Li Caiwen, et al. Preliminary discussion on the phytoplankton assemblage and its response to the environmental changes in the Changjiang (Yangtze) River Estuary and its adjacent waters during the dry season and the wet season[J]. Acta Oceanologica Sinica, 2017, 39(10): 124-144. |
| [161] |
Kong F, Xu Z, Yu R, et al. Distribution patterns of phytoplankton in the Changjiang River Estuary and adjacent waters in spring 2009[J]. Chinese Journal of Oceanology and Limnology, 2016, 34(5): 902-914. |
| [162] |
Li C, Song S, Liu Y, et al. Occurrence of Amoebophrya spp. infection in planktonic dinoflagellates in Changjiang (Yangtze River) Estuary, China[J]. Harmful Algae, 2014, 37: 117-124. |
| [163] |
Liu Y, Song S, Chen T, et al. The diversity and structure of marine protists in the coastal waters of China revealed by morphological observation and 454 pyrosequencing[J]. Estuarine, Coastal and Shelf Science, 2017, 189: 143-155. |
| [164] |
Lu Z, Gan J. Controls of seasonal variability of phytoplankton blooms in the Pearl River Estuary[J]. Deep Sea Research Part Ⅱ: Topical Studies in Oceanography, 2015, 117: 86-96. |
| [165] |
戴明, 李纯厚, 贾晓平, 等. 珠江口近海浮游植物生态特征研究[J]. 应用生态学报, 2004, 15(8): 1389-1394. Dai Ming, Li Chunhou, Jia Xiaoping, et al. Ecological characteristics of phytoplankton in coastal area of Pearl River estuary[J]. Chinese Journal of Applied Ecology, 2004, 15(8): 1389-1394. |
| [166] |
Dong Y, Cui L, Cao R, et al. Ecological characteristics and teratogenic retinal determination of Cochlodinium geminatum blooms in Pearl River Estuary, South China[J]. Ecotoxicology and Environmental Safety, 2020, 191: 110226. |
| [167] |
Shen P P, Li Y N, Qi Y Z, et al. Morphology and bloom dynamics of Cochlodinium geminatum (Schütt) Schütt in the Pearl River Estuary, South China Sea[J]. Harmful Algae, 2012, 13: 10-19. |
| [168] |
黄长江, 董巧香. 1998年春季珠江口海域大规模赤潮原因生物的形态分类和生物学特征Ⅲ[J]. 海洋与湖沼, 2001, 32(1): 1-6. Huang Changjiang, Dong Qiaoxiang. Taxonomic and biological studies on organisms causing a large scale red tide in Zhujiang River Estuary in spring, 1998 Ⅲ[J]. Oceanologia et Limnologia Sinica, 2001, 32(1): 1-6. |
| [169] |
黄长江, 董巧香. 1998年春季珠江口海域大规模赤潮原因生物的形态分类和生物学特征Ⅰ[J]. 海洋与湖沼, 2000, 31(2): 197-204. Huang Changjiang, Dong Qiaoxiang. Taxonomic and biological studies on organisms causing a large scale red tide in Zhujiang River Estuary in spring, 1998Ⅰ[J]. Oceanologia et Limnologia Sinica, 2000, 31(2): 197-204. |
| [170] |
黄长江, 董巧香. 1998年春季珠江口海域大规模赤潮原因生物的形态分类和生物学特征Ⅱ[J]. 海洋与湖沼, 2000, 31(3): 232-238. Huang Changjiang, Dong Qiaoxiang. Taxonomic and biological studies on organisms causing a large scale red tide in Zhujiang River Estuary in spring, 1998Ⅱ[J]. Oceanologia et Limnologia Sinica, 2000, 31(3): 232-238. |
| [171] |
Huang L, Jian W, Song X, et al. Species diversity and distribution for phytoplankton of the Pearl River estuary during rainy and dry seasons[J]. Marine Pollution Bulletin, 2004, 49(7-8): 588-596. |
| [172] |
Qiu D, Huang L, Zhang J, et al. Phytoplankton dynamics in and near the highly eutrophic Pearl River Estuary, South China Sea[J]. Continental Shelf Research, 2010, 30(2): 177-186. |
| [173] |
Li Y, Huang L, Chen J, et al. Water quality and phytoplankton blooms in the Pearl River Estuary[M]. In: Eric W (Ed.), The environment in Asia Pacific harbours. Springer Nature Customer Service Center GmbH, Heidelberg, Germany, 2006: 139-145.
|
| [174] |
Qiu D, Zhong Y, Chen Y, et al. Short‐Term phytoplankton dynamics during typhoon season in and near the Pearl River Estuary, South China Sea[J]. Journal of Geophysical Research: Biogeosciences, 2019, 124(2): 274-292. |
| [175] |
Zhang X, Zhang J, Huang X, et al. Phytoplankton assemblage structure shaped by key environmental variables in the Pearl River Estuary, South China[J]. Journal of Ocean University of China, 2014, 13(1): 73-82. |
| [176] |
Wu W, Liu H. Disentangling protist communities identified from DNA and RNA surveys in the Pearl River–South China Sea continuum during the wet and dry seasons[J]. Molecular Ecology, 2018, 27(22): 4627-4640. |
| [177] |
刘永健, 杨官品, 管晓菁, 等. 胶州湾浮游植物遗传多样性及其季节变化研究[J]. 海洋学报, 2005, 27(5): 103-111. Liu Yongjian, Yang Guanpin, Guan Xiaojing, et al. Study on the genetic diversity and its seasonal variation of phytoplankton in the Jiaozhou Bay[J]. Acta Oceanologica Sinica, 2005, 27(5): 103-111. |
| [178] |
John U, Litaker R W, Montresor M, et al. Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: the introduction of five species with emphasis on molecular-based (rDNA) classification[J]. Protist, 2014, 165(6): 779-804. |
| [179] |
孙军, 刘东艳. 多样性指数在海洋浮游植物研究中的应用[J]. 海洋学报, 2004, 26(1): 62-75. Sun Jun, Liu Dongyan. The application of diversity indices in marine phytoplankton studies[J]. Acta Oceanologica Sinica, 2004, 26(1): 62-75. |
| [180] |
Chen T, Liu Y, Song S, et al. Characterization of the parasitic dinoflagellate Amoebophrya sp. infecting Akashiwo sanguinea in coastal waters of China[J]. Journal Eukaryotic Microbiology, 2018, 65(4): 448-457. |
| [181] |
Berdjeb L, Parada A, Needham D M, et al. Short-term dynamics and interactions of marine protist communities during the spring-summer transition[J]. ISME Journal, 2018, 12(8): 1907-1917. |
| [182] |
Leray M, Knowlton N. Censusing marine eukaryotic diversity in the twenty-first century[J]. Philosophical Transactions of the Royal Society B Biological Sciences, 2016, 371: 20150331. |
| [183] |
胡晓坤, 张清春, 陈振帆, 等. 北部湾海域球形棕囊藻遗传多样性分析[J]. 海洋与湖沼, 2019, 50(3): 601-610. Hu Xiaokun, Zhang Qingchun, Chen Zhenfan, et al. Genetic diversity of Phaeocystis globosa strains iolated from the Beibu Gulf, the South China Sea[J]. Oceanologia et Limnologia Sinica, 2019, 50(3): 601-610. |
| [184] |
Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools[J]. Nucleic Acids Research, 2012, 41(D1): D590-D596. |
| [185] |
Guillou L, Bachar D, Audic S, et al. The protist ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy[J]. Nucleic Acids Research, 2012, 41(D1): D597-D604. |
 2020, Vol. 44
2020, Vol. 44